6G27
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 5 | | Descriptor: | 5-methyl-6-phenyl-2-piperidin-4-yl-pyridazin-3-one, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
2X70
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
4TMV
 
 | |
6I8X
 
 | | As-p18, an extracellular fatty acid binding protein | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein homolog, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Gabrielsen, M, Riboldi-Tunnicliffe, A, Ibanez-Shimabukuro, M, Smith, B.O. | | Deposit date: | 2018-11-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
5E7N
 
 | | Crystal Structure of RPA70N in complex with VU0085636 | | Descriptor: | 2-({3-[(4-bromophenyl)sulfamoyl]-4-methylbenzoyl}amino)benzoic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Gilston, B.A, Patrone, J.D, Pelz, N.F, Bates, B.S, Souza-Fagundes, E.M, Vangamudi, B, Camper, D, Kuznetsov, A, Browning, C.F, Feldkamp, M.D, Olejniczak, E.T, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A.
Chemmedchem, 11, 2016
|
|
2PUG
 
 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
6I6R
 
 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},6~{S})-6-azanyl-2,3,4-tris(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
2WJ2
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(5-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(5-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
1CQZ
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5ZKQ
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6NZB
 
 | |
6K6S
 
 | |
6G29
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 6 | | Descriptor: | 5-methyl-2-piperidin-4-yl-6-pyridin-4-yl-pyridazin-3-one, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2E
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 13 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
5V8M
 
 | | BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
1CG1
 
 | | STRUCTURE OF THE MUTANT (K16Q) OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH HADACIDIN, GDP, 6-PHOSPHORYL-IMP, AND MG2+ | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Choe, J.Y, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-03-26 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic implications from crystalline complexes of wild-type and mutant adenylosuccinate synthetases from Escherichia coli.
Biochemistry, 38, 1999
|
|
6FLO
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei at 2.1 Angstrom resolution | | Descriptor: | GLYCEROL, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-01-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.13868666 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
1CGQ
 
 | |
5A7P
 
 | | Crystal structure of human JMJD2A in complex with compound 36 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6GIW
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-a | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
7X8X
 
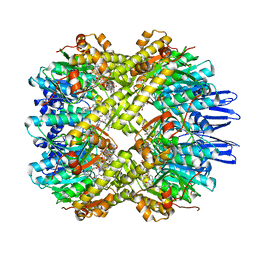 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
6BFG
 
 | |
6GVX
 
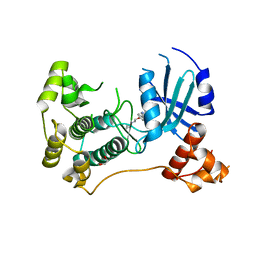 | | Crystal structure of Maternal Embryonic Leucine Zipper Kinase (MELK) in complex with dorsomorphin (Compound C) | | Descriptor: | 1,2-ETHANEDIOL, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Maternal embryonic leucine zipper kinase | | Authors: | Golik, P, Rembacz, K.P, Zrubek, K, Romanowska, M, Bugusz, J, Wladyka, B, Dubin, G. | | Deposit date: | 2018-06-21 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of Maternal Embryonic Leucine Zipper Kinase (MELK) in complex with dorsomorphin (Compound C).
Arch.Biochem.Biophys., 671, 2019
|
|
4QQ8
 
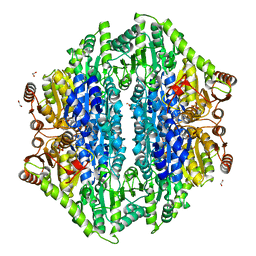 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
