8B9H
 
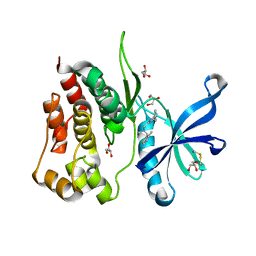 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BIP
 
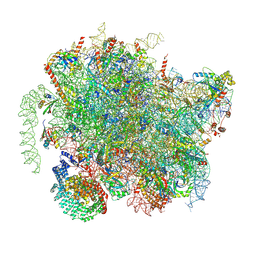 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8B9E
 
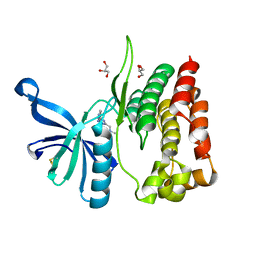 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BBW
 
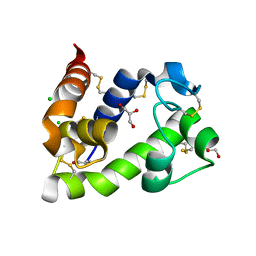 | | Crystal structure of medical leech destabilase (low salt) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into thrombolytic activity of destabilase from medicinal leech.
Sci Rep, 13, 2023
|
|
8B0Q
 
 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
8B9R
 
 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | Descriptor: | Amyloid-beta A4 protein, COPPER (II) ION | | Authors: | Abelein, A, Ciofi-Baffoni, S, Morman, C, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8AUW
 
 | |
8BEW
 
 | |
8AY0
 
 | | Crystal Structure of the peptide binding protein DppE from Bacillus subtilis in complex with murein tripeptide | | Descriptor: | 1,2-ETHANEDIOL, Dipeptide-binding protein DppE, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Hughes, A.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8ATU
 
 | | Cryo-EM structure of human BIRC6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6, ZINC ION | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8AUK
 
 | | Cryo-EM structure of human BIRC6 in complex with HTRA2. | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial, ... | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8BJQ
 
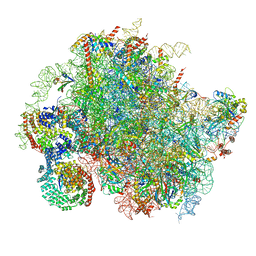 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8B58
 
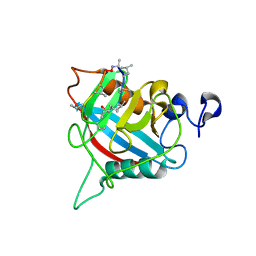 | |
8BFR
 
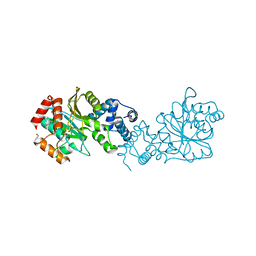 | |
8AUG
 
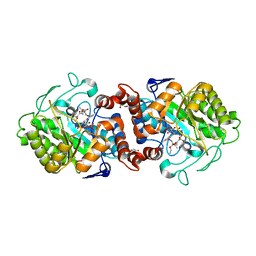 | |
8AX2
 
 | |
8AXT
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | CALCIUM ION, CHLORIDE ION, Sialidase domain-containing protein | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BDT
 
 | | Ternary complex between VCB, BRD4-BD2 and PROTAC 51 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-3-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethylamino]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A.A. | | Deposit date: | 2022-10-20 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
