2RCQ
 
 | | Crystal structure of human apo Cellular Retinol Binding Protein II (CRBP-II) | | Descriptor: | L(+)-TARTARIC ACID, Retinol-binding protein II, cellular, ... | | Authors: | Monaco, H.L, Capaldi, S, Perduca, M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human cellular retinol-binding protein II to 1.2 A resolution.
Proteins, 70, 2007
|
|
2RGD
 
 | | Crystal structure of H-RasQ61L-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Buhrman, G, Wink, G, Mattos, C. | | Deposit date: | 2007-10-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf.
Structure, 15, 2007
|
|
2RIV
 
 | | Crystal structure of the reactive loop cleaved human Thyroxine Binding Globulin | | Descriptor: | GLYCEROL, SULFATE ION, Thyroxine-binding globulin | | Authors: | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | Deposit date: | 2007-10-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
7UOR
 
 | | Crystal structure of cytochrome P450 enzyme CYP119 in complex with methyliridium(III) mesoporphyrin. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome, NICKEL (II) ION, ... | | Authors: | Pereira, J.H, Bloomer, B.J, Hartwig, J.F, Adams, P.D. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Mechanistic and structural characterization of an iridium-containing cytochrome reveals kinetically relevant cofactor dynamics
Nat Catal, 6, 2023
|
|
2RKZ
 
 | |
2RLQ
 
 | | NMR structure of CCP modules 2-3 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-29 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
2RNA
 
 | | Itk SH3 average minimized | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Severin, A.J. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: murine Itk SH3 domain
To be Published
|
|
2R56
 
 | | Crystal Structure of a Recombinant IgE Fab Fragment in Complex with Bovine Beta-Lactoglobulin Allergen | | Descriptor: | Beta-lactoglobulin, DODECYL-BETA-D-MALTOSIDE, IgE Fab Fragment, ... | | Authors: | Niemi, M, Kallio, J.M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen.
Structure, 15, 2007
|
|
7UXF
 
 | | Cryogenic electron microscopy 3D map of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
2R9B
 
 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
7UAH
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7UQU
 
 | |
2RD4
 
 | | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 1, Phospholipase A2 isoform 2, ... | | Authors: | Mirza, Z, Kaur, A, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
To be Published
|
|
7UYO
 
 | |
7UYN
 
 | |
7UYP
 
 | |
2RNE
 
 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
2R4Y
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: H69V/I114M unliganded | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
7UCS
 
 | |
7UCT
 
 | |
7UD3
 
 | |
7UCV
 
 | |
7UCZ
 
 | |
2TRX
 
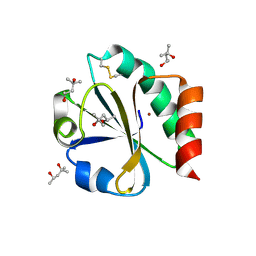 | | CRYSTAL STRUCTURE OF THIOREDOXIN FROM ESCHERICHIA COLI AT 1.68 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, THIOREDOXIN | | Authors: | Katti, S.K, Lemaster, D.M, Eklund, H. | | Deposit date: | 1990-03-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution.
J.Mol.Biol., 212, 1990
|
|
7UCN
 
 | |
