3UQ8
 
 | |
3UQW
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, SULFATE ION, ethyl 1-{(2S,3S)-3-[(3-{[(1R)-1-(4-fluorophenyl)ethyl]carbamoyl}-5-[methyl(methylsulfonyl)amino]benzoyl)amino]-2-hydroxy-4-phenylbutyl}-1H-pyrazole-4-carboxylate | | Authors: | Chen, T.T, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
to be published
|
|
3UYI
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UZX
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and epiandrosterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 3-oxo-5-beta-steroid 4-dehydrogenase, 5-ALPHA-ANDROSTANE-3-BETA,17BETA-DIOL, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
3URR
 
 | |
3V2Q
 
 | | COMPcc in complex with fatty acids | | Descriptor: | Cartilage Oligomerization matrix protein (coiled-coil domain), PALMITIC ACID | | Authors: | Stetefeld, J. | | Deposit date: | 2011-12-12 | | Release date: | 2013-01-16 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentameric channel of COMPcc in complex with different fatty acids.
Plos One, 7, 2012
|
|
3V30
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3UT6
 
 | | Crystal structure of E. Coli PNP complexed with PO4 and formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2011-11-25 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | New phosphate binding sites in the crystal structure of Escherichia coli purine nucleoside phosphorylase complexed with phosphate and formycin A.
Febs Lett., 586, 2012
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3T
 
 | |
3V46
 
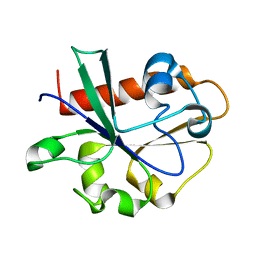 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
3V4F
 
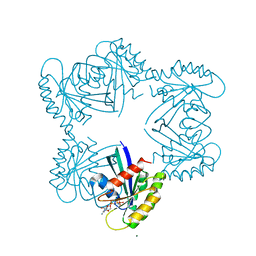 | | H-Ras PEG 400/CaCl2, ordered off | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
3UV4
 
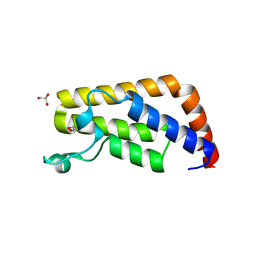 | | Crystal Structure of the second bromodomain of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3V5O
 
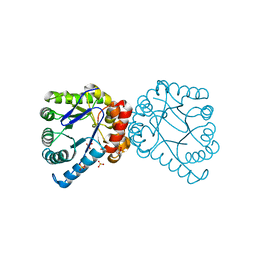 | | Structural and Mechanistic Studies of Catalysis and Sulfa Drug Resistance in Dihydropteroate Synthase | | Descriptor: | Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M, Wu, Y, Li, Z, Zhao, Y, Waddell, M.B, Ferreira, A.M, Lee, R.E, Bashford, D, White, S.W. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3V61
 
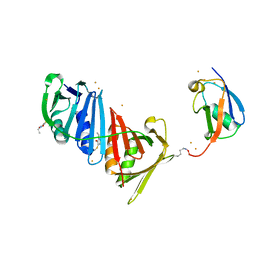 | | Structure of S. cerevisiae PCNA conjugated to SUMO on lysine 164 | | Descriptor: | BARIUM ION, N-ETHYLMALEIMIDE, Proliferating cell nuclear antigen, ... | | Authors: | Armstrong, A.A, Mohideen, F, Lima, C.D. | | Deposit date: | 2011-12-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2.
Nature, 483, 2012
|
|
3V6C
 
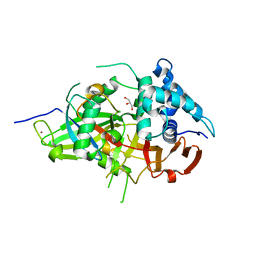 | | Crystal Structure of USP2 in complex with mutated ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of USP2 in complex with mutated ubiquitin
To be Published
|
|
3UWP
 
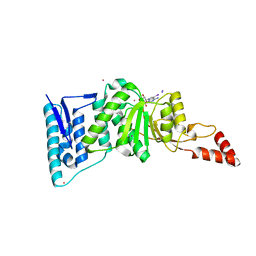 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
3UOA
 
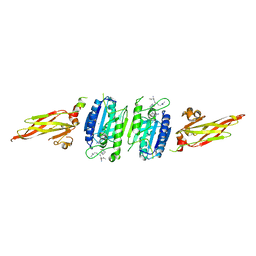 | | Crystal structure of the MALT1 paracaspase (P21 form) | | Descriptor: | MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UP3
 
 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
3UZZ
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and delta4-androstenedione | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
3UPA
 
 | | A general strategy for the generation of human antibody variable domains with increased aggregation resistance | | Descriptor: | kappa light chain variable domain | | Authors: | Dudgeon, K, Rouet, R, Kokmeijer, I, Langley, D.B, Christ, D. | | Deposit date: | 2011-11-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General strategy for the generation of human antibody variable domains with increased aggregation resistance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQ7
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240S F247L (L9S F16L) in presence of 10 mM cysteamine | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V21
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*TP*CP*GP*AP*CP*CP*GP*GP*TP*CP*GP*A)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
3USF
 
 | | Crystal structure of DAVA-4 | | Descriptor: | (4S)-4,5-DIAMINOPENTANOIC ACID, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2011-11-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of DAVA-4
To be Published
|
|
3UTE
 
 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase sulfate complex | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
