6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
5W9K
 
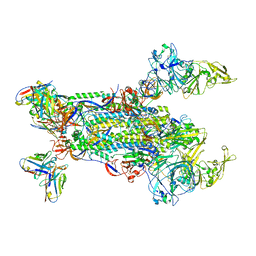 | |
3M19
 
 | |
6ST9
 
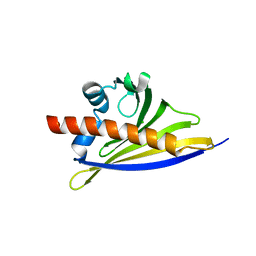 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | Descriptor: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
5JMC
 
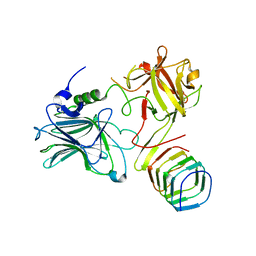 | | Receptor binding domain of Botulinum neurotoxin A in complex with rat SV2C | | Descriptor: | Botulinum neurotoxin type A, Synaptic vesicle glycoprotein 2C | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2YCD
 
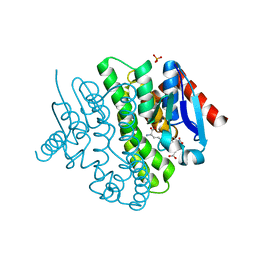 | | Structure of a novel Glutathione Transferase from Agrobacterium tumefaciens. | | Descriptor: | GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, S-(P-NITROBENZYL)GLUTATHIONE | | Authors: | Skopelitou, K, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2011-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Glutathione Transferase from Agrobacterium Tumefaciens Reveals a Novel Class of Bacterial Gst Superfamily.
Plos One, 7, 2012
|
|
5NHV
 
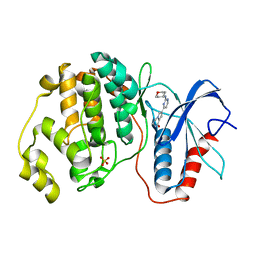 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6RX7
 
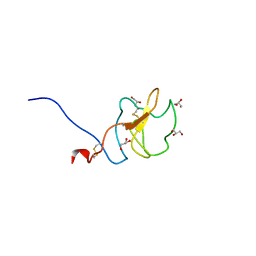 | |
9GY6
 
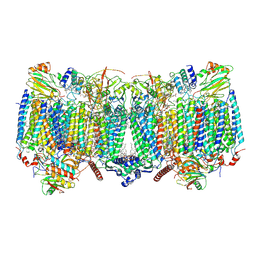 | | Mycobacterial cytochrome bc1:aa3 with inhibitor | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[2-(trifluoromethylsulfonyl)-2-azaspiro[3.3]heptan-6-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | lamers, M.H, Verma, A.K. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic study of a novel inhibitor analogue of M. tuberculosis cytochrome bc 1 :aa 3 .
NPJ Drug Discov, 2, 2025
|
|
6RFW
 
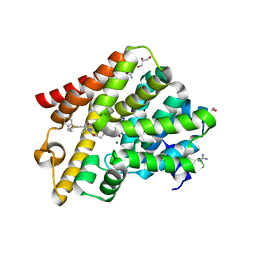 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1039 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-[2-(2-fluorophenyl)ethyl]prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
5L8C
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5O4A
 
 | | Human FGF in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, SULFATE ION, ... | | Authors: | Debreczeni, J, Breed, J, Mukherjee, H, Aquila, B, Kaiser, C, Tentarelli, S, Whitty, A, Grimster, N. | | Deposit date: | 2017-05-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A study of the reactivity of S(VI)-F containing warheads with nucleophilic amino-acid side chains under physiological conditions.
Org. Biomol. Chem., 15, 2017
|
|
3N4N
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S. | | Deposit date: | 2010-05-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
6KDX
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]quinolin-2-amine, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6EIT
 
 | |
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
6CGM
 
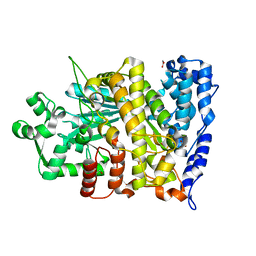 | |
5JNP
 
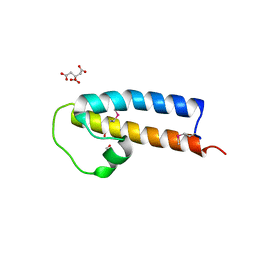 | | Crystal structure of a rice (Oryza Sativa) cellulose synthase plant conserved region (P-CR) | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Rushton, P.S, Olek, A.T, Makowski, L, Badger, J, Steussy, C.N, Carpita, N.C, Stauffacher, C.V. | | Deposit date: | 2016-04-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Rice Cellulose SynthaseA8 Plant-Conserved Region Is a Coiled-Coil at the Catalytic Core Entrance.
Plant Physiol., 173, 2017
|
|
5B60
 
 | | Crystal structure of PtLCIB4 S47R mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | CHLORIDE ION, PtLCIB4 S47R mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6CPG
 
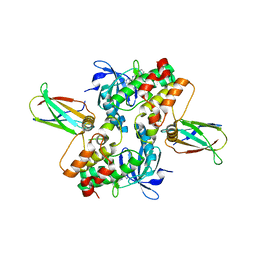 | | Structure of dephosphorylated Aurora A (122-403) in complex with inhibiting monobody and AT9283 in an inactive conformation | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Aurora kinase A, Monobody | | Authors: | Otten, R, Kutter, S, Zorba, A, Padua, R.A.P, Koide, A, Koide, S, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
6CGL
 
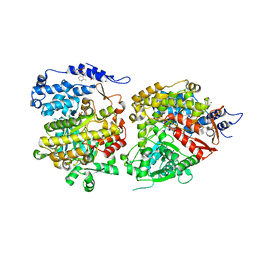 | |
5JLV
 
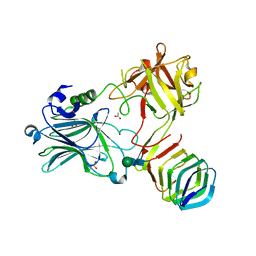 | | Receptor binding domain of Botulinum neurotoxin A in complex with human glycosylated SV2C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6KDZ
 
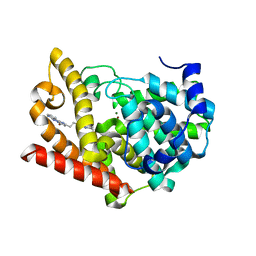 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | 4-[2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-3,7,8,10-tetrazatricyclo[7.4.0.0^{2,7}]trideca-1,3,5,8,10,12-hexaen-6-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
5B5Z
 
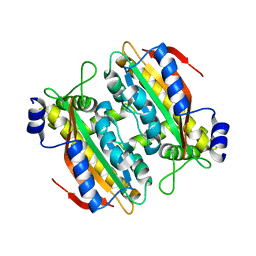 | | Crystal structure of PtLCIB4 H88A mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | PtLCIB4 H88A mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7MM6
 
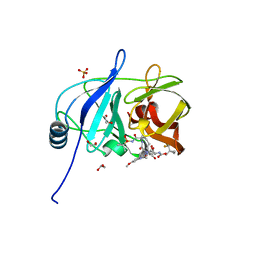 | |
