2WNQ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in space group P21 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
3PVE
 
 | | Crystal structure of the G2 domain of Agrin from Mus Musculus | | Descriptor: | Agrin, Agrin protein | | Authors: | Sampathkumar, P, Do, J, Bain, K, Freeman, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the G2 domain of Agrin from Mus Musculus
To be Published
|
|
4HC8
 
 | |
3NZ7
 
 | |
4DO2
 
 | |
2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WPB
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate and the inhibitor (2R,3R)-2,3,4- trihydroxy-N,N-dipropylbutanamide in space group P21 crystal form I | | Descriptor: | (2R,3R)-2,3,4-TRIHYDROXY-N,N-DIPROPYLBUTANAMIDE, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
4JWT
 
 | |
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
4IPC
 
 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
3OPH
 
 | | ESBL R164S mutant of SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sampson, J.M, van den Akker, F. | | Deposit date: | 2010-09-01 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase.
Antimicrob.Agents Chemother., 55, 2011
|
|
4JP2
 
 | | Crystal Structure of TT0495 protein from Thermus thermophilus HB8 | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase | | Authors: | Pampa, K.J, Lokanath, N.K, Kunishima, N, Ravishnkar Rai, V. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structure of NAD-dependent 3-dehydro-2-deoxy-D-gluconate dehydrogenase from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
4JOS
 
 | | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335) | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenosylhomocysteine nucleosidase, ... | | Authors: | Sampathkumar, P, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335)
to be published
|
|
4J8R
 
 | | Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment | | Descriptor: | Heavy chain of POM2 Fab, Light chain of POM2 Fab, Major prion protein | | Authors: | Swayampakula, M, Baral, P.K, Kav, N.N.V, Aguzzi, A, James, M.N.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The crystal structure of an octapeptide repeat of the Prion protein in complex with a Fab fragment of the POM2 antibody.
Protein Sci., 22, 2013
|
|
3H14
 
 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3K16
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
4IFQ
 
 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
3H9M
 
 | | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, TRIETHYLENE GLYCOL, p-aminobenzoate synthetase, ... | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Do, J, Bain, K, Rutter, M, Gheyi, T, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii
To be Published
|
|
3NO8
 
 | | Crystal structure of the PHR domain from human BTBD2 Protein | | Descriptor: | BTB/POZ domain-containing protein 2, GLYCEROL, SULFATE ION | | Authors: | Sampathkumar, P, Miller, S, Rutter, M, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the PHR domain from human BTBD2 Protein
To be Published
|
|
3NWA
 
 | | Glycoprotein B from Herpes simplex virus type 1, W174R mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2629 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NWD
 
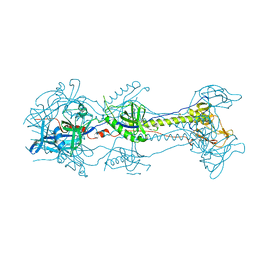 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8803 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
4GGG
 
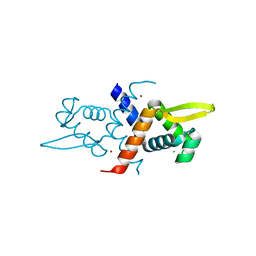 | | Crystal structure of V66A/L68V CzrA in the Zn(II)bound state. | | Descriptor: | CHLORIDE ION, Repressor protein, ZINC ION | | Authors: | Campanello, G.C, Ma, Z, Grossoehme, N.E, Chakrovorty, D.K, Guerra, A.J, Ye, Y, Dann III, C.E, Merz Jr, K.M, Giedroc, D.P. | | Deposit date: | 2012-08-06 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Allosteric inhibition of a zinc-sensing transcriptional repressor: insights into the arsenic repressor (ArsR) family.
J.Mol.Biol., 425, 2013
|
|
