7S5P
 
 | |
6N5X
 
 | | Crystal structure of the SNX5 PX domain in complex with the CI-MPR (space group P212121 - Form 1) | | Descriptor: | GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Sorting nexin-5,Cation-independent mannose-6-phosphate receptor | | Authors: | Collins, B, Paul, B, Weeratunga, S. | | Deposit date: | 2018-11-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins.
Nat.Cell Biol., 21, 2019
|
|
6MWE
 
 | | CRYSTAL STRUCTURE OF TIE2 IN COMPLEX WITH DECIPERA COMPOUND DP1919 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Angiopoietin-1 receptor, SULFATE ION | | Authors: | Chun, L, Abendroth, J, Edwards, T.E. | | Deposit date: | 2018-10-29 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Selective Tie2 Inhibitor Rebastinib Blocks Recruitment and Function of Tie2
Mol. Cancer Ther., 16, 2017
|
|
6FR2
 
 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
6FTN
 
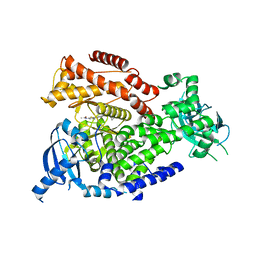 | | mPI3Kd IN COMPLEX WITH AZ2 | | Descriptor: | Phosphor inositol 3 kinase, ~{N}-[5-[2-[(1~{S})-1-cyclopropylethyl]-7-methyl-1-oxidanylidene-3~{H}-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Highly Isoform Selective Orally Bioavailable Phosphoinositide 3-Kinase (PI3K)-gamma Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FWY
 
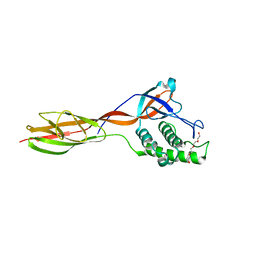 | |
6N4F
 
 | |
6FX6
 
 | |
6N5Z
 
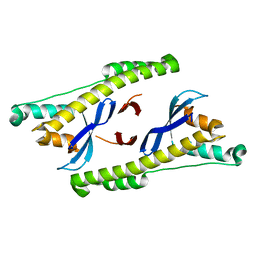 | |
6G3X
 
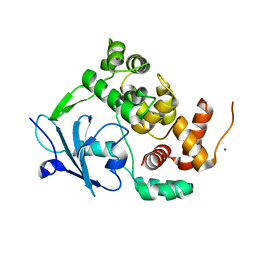 | |
6MZB
 
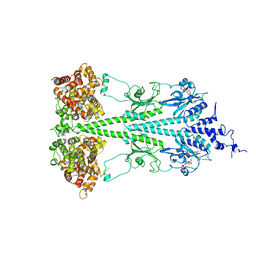 | | Cryo-EM structure of phosphodiesterase 6 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Gulati, S, Palczewski, K. | | Deposit date: | 2018-11-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.
Sci Adv, 5, 2019
|
|
6N5Y
 
 | |
6FVR
 
 | |
7SA6
 
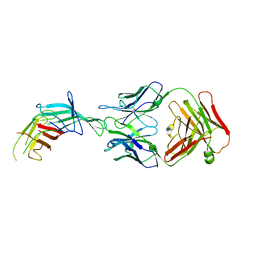 | | fHbp mutant 2416 bound to Fab JAR5 | | Descriptor: | Factor H-binding protein 2416, JAR5 Heavy Chain, JAR5 Light Chain | | Authors: | Chesterman, C, Malito, E, Bottomley, M.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active Learning for Rapid Design: An iterative AI approach for accelerated vaccine design that combines active machine learning and high-throughput experimental evaluation
To Be Published
|
|
7S4E
 
 | |
7SBZ
 
 | |
6FLR
 
 | | Super-open structure of the AMPAR GluA3 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3 | | Authors: | Garcia-Nafria, J. | | Deposit date: | 2018-01-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
6FN1
 
 | | Zosuquidar and UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human-mouse chimeric ABCB1 (ABCBHM), UIC2 Antigen Binding Fragment Heavy Chain, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FVS
 
 | |
6G2M
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor PB-PAU | | Descriptor: | 1,2-ETHANEDIOL, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2018-03-23 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-based optimization of bisphosphonate nucleoside inhibitors of human 5'(3')-deoxyribonucleotidases
Eur.J.Org.Chem., 2018
|
|
6FTW
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-048 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}prop-2-ynamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-24 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6G3Y
 
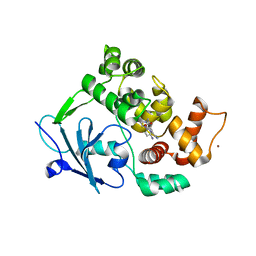 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
6NSV
 
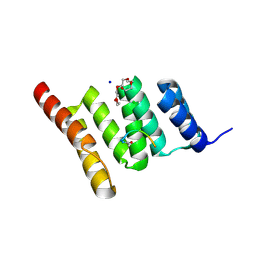 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
7S9N
 
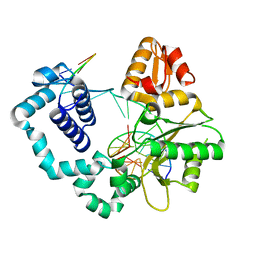 | | Binary complex of DNA Polymerase Beta with Fapy-dG in the template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(FAP)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J, Smith, M.R. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
7S9J
 
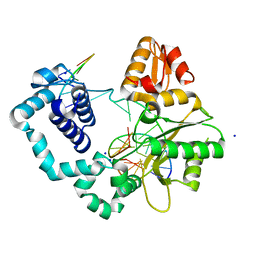 | | Crystal Structure of DNA Polymerase Beta with Fapy-dG base-paired with a dC | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*(FAP)P*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
