6ME4
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
8IBI
 
 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
6S3A
 
 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, CHLORIDE ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-06-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
6OMT
 
 | | HIV-1 capsid hexamer R18D mutant | | Descriptor: | Capsid protein | | Authors: | Huang, P, Summers, B.J, Xiong, Y. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-21 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | FEZ1 Is Recruited to a Conserved Cofactor Site on Capsid to Promote HIV-1 Trafficking.
Cell Rep, 28, 2019
|
|
5I56
 
 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
6EMW
 
 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
7N60
 
 | | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lau, I.C.K, Coleman, T, Bell, S.G, Bruning, J.B. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant
To Be Published
|
|
5I63
 
 | |
1BC0
 
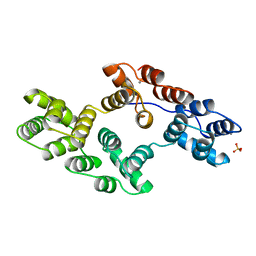 | | RECOMBINANT RAT ANNEXIN V, W185A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
8IAN
 
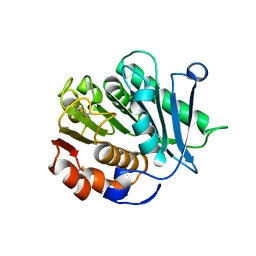 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
7B0G
 
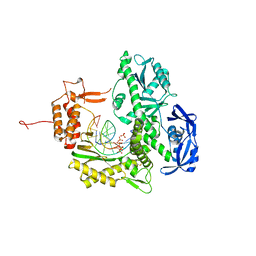 | | TgoT_6G12 binary with 2 hCTPs | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*TP*TP*GP*GP*CP*TP*GP*CP*CP*CP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*GP*CP*AP*GP*()P*())-3'), ... | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7POA
 
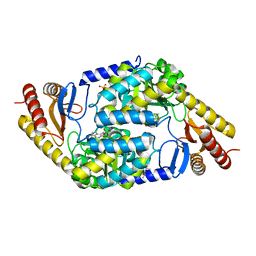 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POB
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POC
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
1AU2
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PROPANONE INHIBITOR | | Descriptor: | 1-[[(4-PHENOXYPHENYL)SULFONYL]AMINO]-3-[[N/N-(4-PYRIDINYLCARBONYL)-L-LEUCYL]AMINO]-2-PROPANOL, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
5IDE
 
 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model I) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
5E6G
 
 | |
7N3Z
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
8DFR
 
 | | REFINED CRYSTAL STRUCTURES OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE. 3 ANGSTROMS APO-ENZYME AND 1.7 ANGSTROMS NADPH HOLO-ENZYME COMPLEX | | Descriptor: | CALCIUM ION, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Matthews, D.A, Oatley, S.J, Burridge, J, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined Crystal Structures of Chicken Liver Dihydrofolate Reductase. 3 Angstroms Apo-Enzyme and 1.7 Angstroms Nadph Holo-Enzyme Complex
To be Published
|
|
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
6YI0
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.65 A in P41212 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, SODIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
8DII
 
 | |
6J8L
 
 | |
5DXT
 
 | | p110alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, 1,2-ETHANEDIOL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DYH
 
 | | Ti(IV) bound human serum transferrin | | Descriptor: | CARBONATE ION, CITRIC ACID, Serotransferrin, ... | | Authors: | Saxena, M, Sharma, S, Noinaj, N, Parks, T.B, Tinoco, A.D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Unusual Synergism of Transferrin and Citrate in the Regulation of Ti(IV) Speciation, Transport, and Toxicity.
J.Am.Chem.Soc., 138, 2016
|
|
