4L16
 
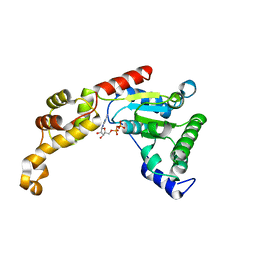 | | Crystal structure of FIGL-1 AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
3NOQ
 
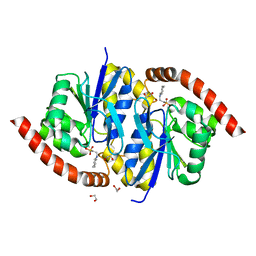 | | Crystal Structure of C101S Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
2PK6
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10033 | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl- L-cysteinyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
3BET
 
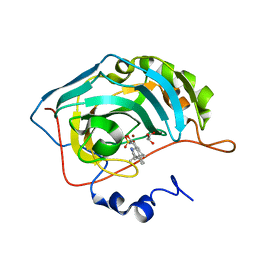 | | Crystal structure of the human carbonic anhydrase II in complex with STX 641 at 1.85 angstroms resolution | | Descriptor: | (17beta)-17-(cyanomethyl)-2-methoxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2007-11-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-activity relationships of C-17 cyano-substituted estratrienes as anticancer agents
J.Med.Chem., 51, 2008
|
|
5GJW
 
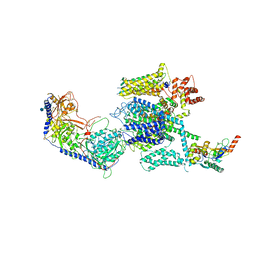 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex for ClassII map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
3VPE
 
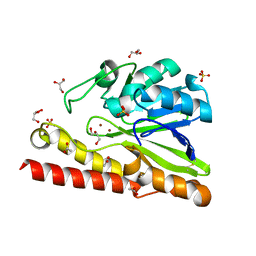 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 | | Descriptor: | ACETATE ION, GLYCEROL, Metallo-beta-lactamase, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
4L7J
 
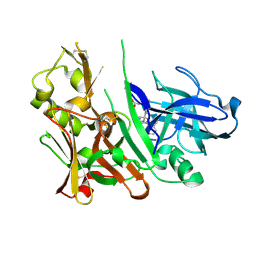 | | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxy-benzylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against Beta-Secretase-1 (BACE-1) | | Descriptor: | 2-[(3aS,7aR)-2-amino-3a-(2,4-difluorophenyl)-3a,6,7,7a-tetrahydro[1,3]oxazolo[4,5-c]pyridin-5(4H)-yl]pyridine-3-carbonitrile, ACETATE ION, Beta-secretase 1 | | Authors: | Huestis, M.P, Liu, W, Volgraf, M, Purkey, H.E, Wang, W, Yu, C, Wu, P, Smith, D, Vigers, G, Dutcher, D, Geck Do, M.K, Hunt, K.W, Siu, M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxycycloalkylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against β-Secretase-1 (BACE-1)
Tetrahedron Lett., 2013
|
|
2WJ1
 
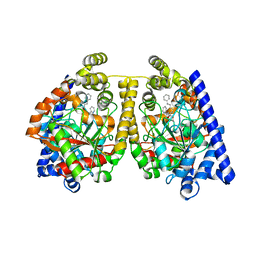 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
4CNH
 
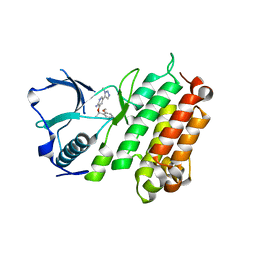 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy)-5-(1-methyl-1H- 1,2,3-triazol-5-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy]-5-(1-methyl-1H-1,2,3-triazol-5-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
7KVQ
 
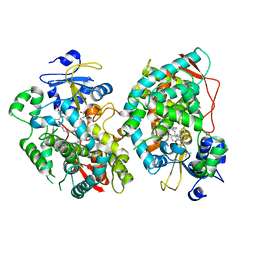 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
3WKG
 
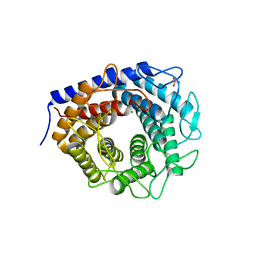 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3EOX
 
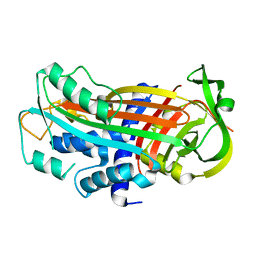 | |
2HUU
 
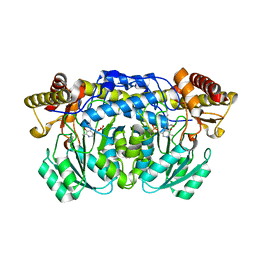 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with alanine | | Descriptor: | 1-BUTANOL, ALANINE, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
4H0D
 
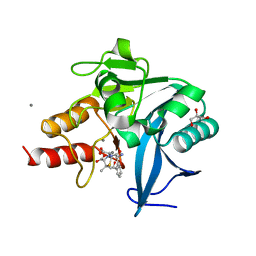 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae
To be Published
|
|
3WKI
 
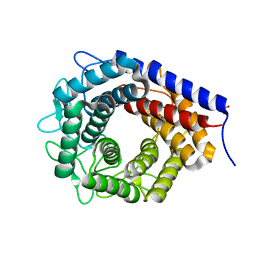 | | Crystal structure of cellobiose 2-epimerase in complex with cellobiitol | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
2HVM
 
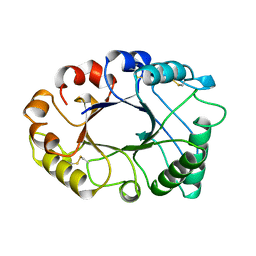 | |
1F4Z
 
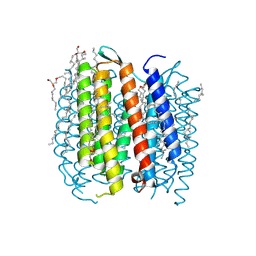 | | BACTERIORHODOPSIN-M PHOTOINTERMEDIATE STATE OF THE E204Q MUTANT AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
1F58
 
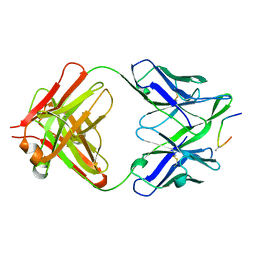 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
1W1D
 
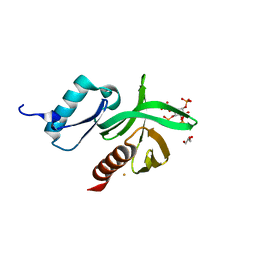 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, GOLD ION, ... | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
2I4Q
 
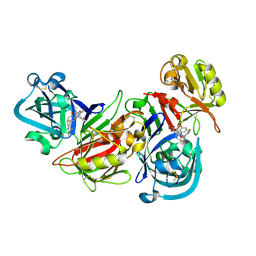 | | Human renin/PF02342674 complex | | Descriptor: | (2S)-6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2-(3,5-DIFLUOROPHENYL)-4-(3-METHOXYPROPYL)-2-METHYL-2H-1,4-BENZOXAZIN-3(4H)-ONE, Renin | | Authors: | Holsworth, D.D, Jalaie, M, Zhang, E, Mcconnell, P, Mochalkin, I, Finzel, B.C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of 6-(2,4-diaminopyrimidinyl)-1,4-benzoxazin-3-ones as small molecule renin inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
1F50
 
 | | BACTERIORHODOPSIN-BR STATE OF THE E204Q MUTANT AT 1.7 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
7EDH
 
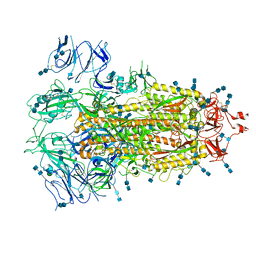 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1F5N
 
 | | HUMAN GUANYLATE BINDING PROTEIN-1 IN COMPLEX WITH THE GTP ANALOGUE, GMPPNP. | | Descriptor: | INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Prakash, B, Renault, L, Praefcke, G.J.K, Herrmann, C, Wittinghofer, A. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism.
EMBO J., 19, 2000
|
|
7EDF
 
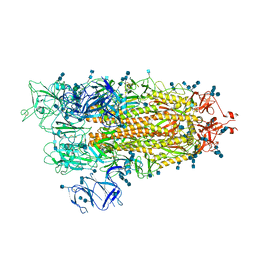 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDG
 
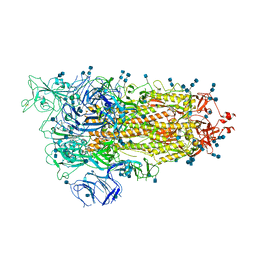 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
