3R44
 
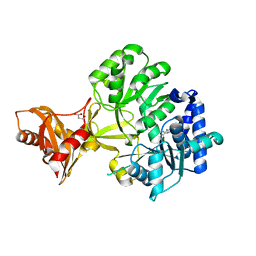 | | Mycobacterium tuberculosis fatty acyl CoA synthetase | | Descriptor: | HISTIDINE, MALONATE ION, fatty acyl CoA synthetase FADD13 (FATTY-ACYL-CoA SYNTHETASE) | | Authors: | Andersson, C.S, Martinez Molina, D, Hogbom, M. | | Deposit date: | 2011-03-17 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme.
Structure, 20, 2012
|
|
8A9B
 
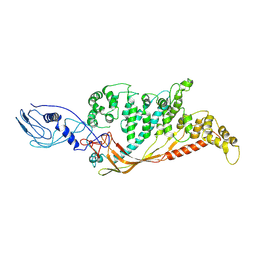 | |
8A9A
 
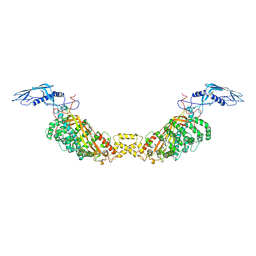 | |
3R5L
 
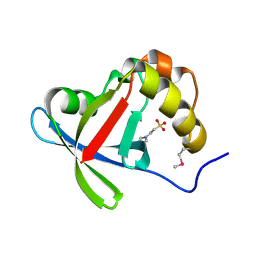 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
7ADK
 
 | | Structure of the mycoplasma MIB and MIP proteins | | Descriptor: | Lipoprotein, Putative immunoglobulin-blocking virulence protein | | Authors: | Nottelet, P, Bataille, L, Gourgues, G, Anger, R, Lartigue, C, Sirand-Pugnet, P, Marza, E, Fronzes, R, Arfi, Y. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction.
Sci Adv, 7, 2021
|
|
3QWG
 
 | |
3R1F
 
 | | Crystal structure of a key regulator of virulence in Mycobacterium tuberculosis | | Descriptor: | ESX-1 secretion-associated regulator EspR | | Authors: | Rosenberg, O.S, Dovey, C, Finer-Moore, J, Stroud, R.M, Cox, J.S. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TZ6
 
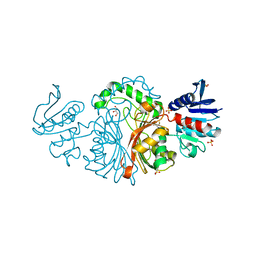 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1C3V
 
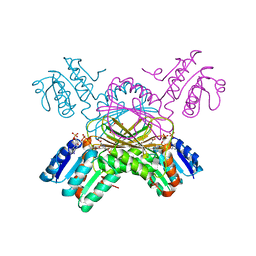 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
3UC5
 
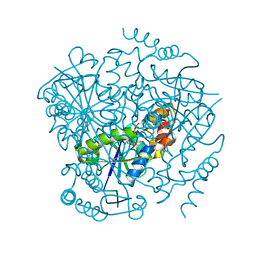 | | Phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphopantetheine adenylyltransferase | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray study of the conformational changes in the molecule of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis during the catalyzed reaction.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7ADJ
 
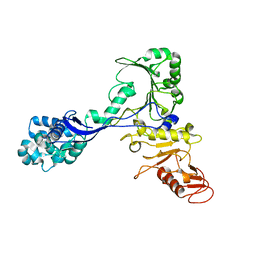 | | Structure of the mycoplasma MIB protein | | Descriptor: | Putative immunoglobulin-blocking virulence protein | | Authors: | Nottelet, P, Bataille, L, Gourgues, G, Anger, R, Lartigue, C, Sirand-Pugnet, P, Marza, E, Fronzes, R, Arfi, Y. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction.
Sci Adv, 7, 2021
|
|
7ADM
 
 | | Structure of the mycoplasma MIB protein | | Descriptor: | Putative immunoglobulin-blocking virulence protein | | Authors: | Nottelet, P, Bataille, L, Gourgues, G, Anger, R, Lartigue, C, Sirand-Pugnet, P, Marza, E, Fronzes, R, Arfi, Y. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction.
Sci Adv, 7, 2021
|
|
7ZTA
 
 | | Structure of an Escherichia coli 70S ribosome stalled by Tetracenomycin X during translation of an MAAAPQK(C) peptide | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Leroy, E.C, Perry, T.N, Renault, T.T, Innis, C.A. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Tetracenomycin X sequesters peptidyl-tRNA during translation of QK motifs.
Nat.Chem.Biol., 19, 2023
|
|
1YIC
 
 | | THE OXIDIZED SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C, ISO-1, HEME C | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1997-02-18 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized Saccharomyces cerevisiae iso-1-cytochrome c.
Biochemistry, 36, 1997
|
|
8DWM
 
 | | Host-guest complex of bleomycin A2 fully bound to CTTAGTTATAACTAAG | | Descriptor: | BLEOMYCIN A2, COBALT (III) ION, DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), ... | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-08-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
6VDD
 
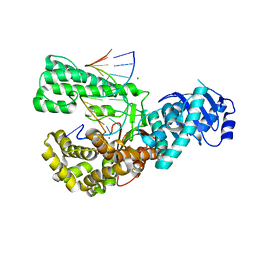 | | POL domain of Pol1 from M. smegmatis complex with DNA primer-template and dNTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A*(DCT))-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDC
 
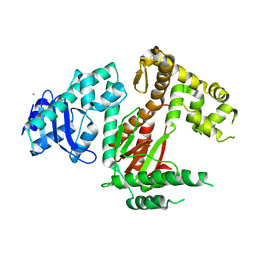 | | POL domain of Pol1 from M. smegmatis | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDE
 
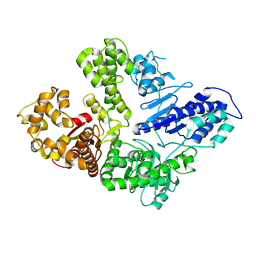 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
8R37
 
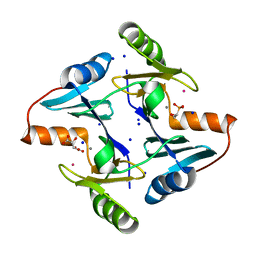 | | Klebsiella pneumoniae fosfomycin-resistance protein (FosAKP) | | Descriptor: | FOSFOMYCIN, FosA family fosfomycin resistance glutathione transferase, L(+)-TARTARIC ACID, ... | | Authors: | Papageorgiou, A.C, Varotsou, C, Labrou, N.E. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Studies of Klebsiella pneumoniae Fosfomycin-Resistance Protein and Its Application for the Development of an Optical Biosensor for Fosfomycin Determination.
Int J Mol Sci, 25, 2023
|
|
8PC1
 
 | |
8PBZ
 
 | |
8PC0
 
 | | Sub-tomogram average of the open conformation of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. | | Descriptor: | Adhesin P1, Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-1,5-anhydro-D-glucitol, ... | | Authors: | Sprankel, L, Scheffer, M.P, Frangakis, A.F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium.
Plos Pathog., 19, 2023
|
|
7F0A
 
 | |
7F0F
 
 | | Crystal structure of capreomycin phosphotransferase in complex with CMN IIB | | Descriptor: | Capreomycin phosphotransferase, DPP-ALA-DPP-UAL-MYN-KBE | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
