1KTR
 
 | | Crystal Structure of the Anti-His Tag Antibody 3D5 Single-Chain Fragment (scFv) in Complex with a Oligohistidine peptide | | Descriptor: | Anti-his tag antibody 3d5 variable light chain, Peptide linker, Anti-his tag antibody 3d5 variable heavy chain, ... | | Authors: | Kaufmann, M, Lindner, P, Honegger, A, Blank, K, Tschopp, M, Capitani, G, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-01-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the anti-His tag antibody 3D5 single-chain fragment complexed to its antigen.
J.Mol.Biol., 318, 2002
|
|
3GA4
 
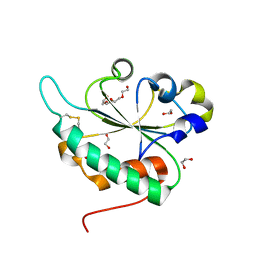 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6SWH
 
 | | Crystal structure of the ternary complex between the type 1 pilus proteins FimC, FimI and FimA from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giese, C, Puorger, C, Ignatov, O, Weber, M, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
3HBX
 
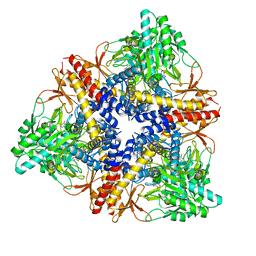 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
2P2C
 
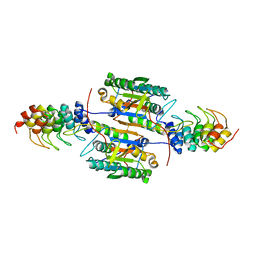 | | Inhibition of caspase-2 by a designed ankyrin repeat protein (DARPin) | | Descriptor: | Caspase-2 | | Authors: | Roschitzki Voser, H, Briand, C, Capitani, G, Gruetter, M.G. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism.
Structure, 15, 2007
|
|
4HU7
 
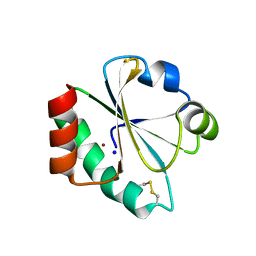 | | E. coli thioredoxin variant with Pro76 as single proline residue | | Descriptor: | COPPER (II) ION, SODIUM ION, Thioredoxin-1 | | Authors: | Glockshuber, R, Scharer, M.A, Capitani, G, Rubini, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
4HUA
 
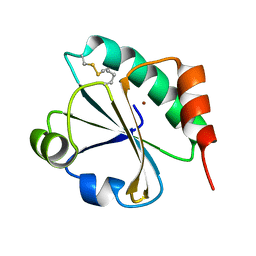 | | E. coli thioredoxin variant with (4R)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
6ERJ
 
 | | Self-complemented FimA subunit from Salmonella enterica | | Descriptor: | ACETIC ACID, Type-1 fimbrial protein, a chain | | Authors: | Zyla, D.S, Prota, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 294, 2019
|
|
6ZRW
 
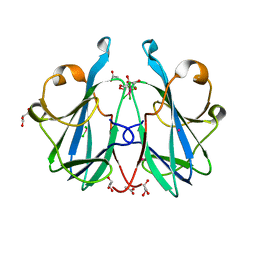 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
4JRA
 
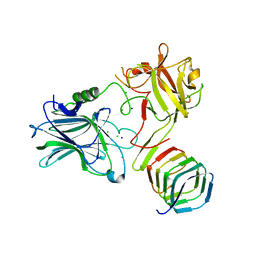 | | CRYSTAL STRUCTURE OF THE BOTULINUM NEUROTOXIN A RECEPTOR-BINDING DOMAIN IN COMPLEX WITH THE LUMINAL DOMAIN Of SV2C | | Descriptor: | Botulinum neurotoxin type A, CHLORIDE ION, SODIUM ION, ... | | Authors: | Benoit, R.M, Frey, D, Wieser, M.M, Jaussi, R, Schertler, G.F.X, Capitani, G, Kammerer, R.A. | | Deposit date: | 2013-03-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A.
Nature, 505, 2014
|
|
4HU9
 
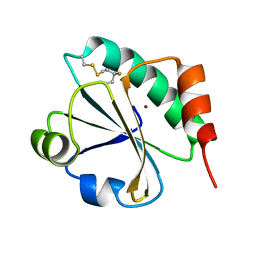 | | E. coli thioredoxin variant with (4S)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
7VCR
 
 | |
7VCN
 
 | |
7W6B
 
 | |
8PC7
 
 | |
6RB0
 
 | |
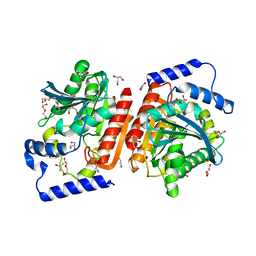
6RKY
 
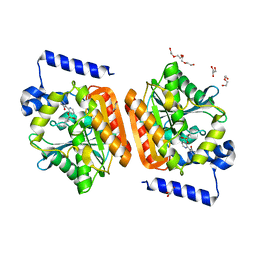 | |
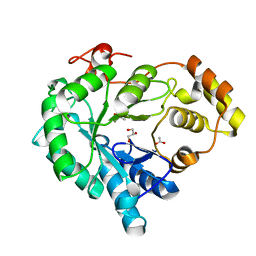
6I8F
 
 | |
6KBL
 
 | | Structure-function study of AKR4C14, an aldo-keto reductase from Thai Jasmine rice (Oryza sativa L. ssp. Indica cv. KDML105) | | Descriptor: | ACETATE ION, Aldo-keto reductase, CACODYLATE ION, ... | | Authors: | Songsiriritthigul, C, Narawongsanont, R, Guan, H.H, Chen, C.J. | | Deposit date: | 2019-06-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function study of AKR4C14, an aldo-keto reductase from Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105).
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8FIZ
 
 | | Cryo-EM structure of E. coli 70S Ribosome containing mRNA and tRNA (in the transcription-translation complex) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIY
 
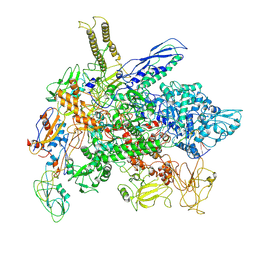 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIX
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
6JLR
 
 | |
4W99
 
 | | Apo-structure of the Y79F,W322E-double mutant of Etr1p | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Erb, T.J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
6NEU
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
