2Y2P
 
 | | Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2L
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E06) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2M
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E08) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2K
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
3QNC
 
 | | Crystal Structure of a Rationally Designed OXA-10 Variant Showing Carbapenemase Activity, OXA-10loop48 | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, Oxacillinase, ... | | Authors: | De Luca, F, Benvenuti, M, Carboni, F, Pozzi, C, Rossolini, G.M, Mangani, S, Docquier, J.D. | | Deposit date: | 2011-02-08 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D {beta}-lactamase OXA-10 by rational protein design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QNB
 
 | | Crystal Structure of an Engineered OXA-10 Variant with Carbapenemase Activity, OXA-10loop24 | | Descriptor: | 1,2-ETHANEDIOL, Oxacillinase, SULFATE ION | | Authors: | De Luca, F, Benvenuti, M, Carboni, F, Pozzi, C, Rossolini, G.M, Mangani, S, Docquier, J.D. | | Deposit date: | 2011-02-08 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D {beta}-lactamase OXA-10 by rational protein design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UE1
 
 | | Crystal strucuture of Acinetobacter baumanni PBP1A in complex with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE3
 
 | | Crystal structure of Acinetobacter baumanni PBP3 | | Descriptor: | Septum formation, penicillin binding protein 3, peptidoglycan synthetase | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDF
 
 | | Crystal structure of Apo PBP1a from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDI
 
 | |
3UE0
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDX
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UY6
 
 | |
3VMA
 
 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Huang, C.Y, Sung, M.T, Lai, Y.T, Chou, L.Y, Shih, H.W, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4DKI
 
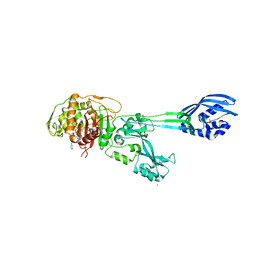 | | Structural Insights into the Anti- Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BICARBONATE ION, CADMIUM ION, ... | | Authors: | Lovering, A.L, Gretes, M.C, Strynadka, N.C.J. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Anti-methicillin-resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole.
J.Biol.Chem., 287, 2012
|
|
4FSF
 
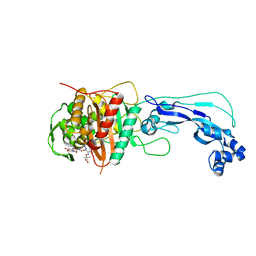 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3UPP
 
 | |
3UPN
 
 | |
3UPO
 
 | |
3UN7
 
 | |
3VSK
 
 | |
3VSL
 
 | | Crystal structure of penicillin-binding protein 3 (PBP3) from methicilin-resistant Staphylococcus aureus in the cefotaxime bound form. | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Yoshida, H, Tame, J.R, Park, S.Y. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein 3 (PBP3) from Methicillin-Resistant Staphylococcus aureus in the Apo and Cefotaxime-Bound Forms.
J.Mol.Biol., 423, 2012
|
|
4BD0
 
 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4BD1
 
 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4JBF
 
 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
