8U69
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-chloro-5-(4-chloro-2-(2-(5-chloro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)benzonitrile (JLJ334), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(5-chloro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6N
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)-N,N-dimethylpropanamide (JLJ752), a non-nucleoside inhibitor | | Descriptor: | 3-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}-N,N-dimethylpropanamide, Gag-Pol polyprotein, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7OZ5
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA in complex with fragment 166 at the transient P-pocket. | | Descriptor: | (1~{R},2~{R})-2-phenyl-~{N}-(1,3-thiazol-2-yl)cyclopropane-1-carboxamide, CADMIUM ION, DNA (28-MER), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
6NFP
 
 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
7SVT
 
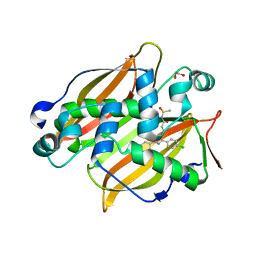 | | Mycobacterium tuberculosis 3-hydroxyl-ACP dehydratase HadAB in complex with 1,3-diarylpyrazolyl-acylsulfonamide inhibitor | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 1,2-ETHANEDIOL, 3-[1-(4-bromophenyl)-3-(4-chlorophenyl)-1H-pyrazol-4-yl]-N-(methanesulfonyl)propanamide, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1,3-Diarylpyrazolyl-acylsulfonamides Target HadAB/BC Complex in Mycobacterium tuberculosis .
Acs Infect Dis., 8, 2022
|
|
8A45
 
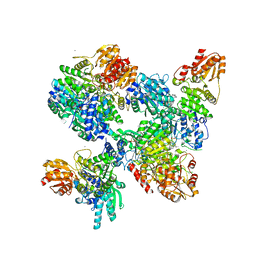 | |
8TDC
 
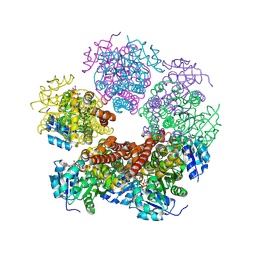 | | Structure of PYCR1 complexed with NADH and 1,3-dithiane-2-carboxylic acid | | Descriptor: | 1,3-dithiane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD4
 
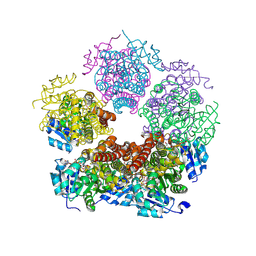 | | Structure of PYCR1 complexed with NADH and 1,3-Dithiolane-2-carboxylic acid | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD2
 
 | | Structure of PYCR1 complexed with NADH and cyclobutane-1,1-dicarboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
6O3G
 
 | |
8A29
 
 | | Apo 1-deoxy-D-xylulose 5-phosphate synthase from Pseudomonas aeruginosa | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hamid, R, Adam, S, Lacour, A, Monjas, L, Hirsch, A. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1-deoxy-D-xylulose-5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding.
J.Biol.Chem., 299, 2023
|
|
8A5K
 
 | |
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6YOZ
 
 | | HiCel7B labelled with b-1,4-glucosyl cyclophellitol | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
6O3L
 
 | |
8QBQ
 
 | | Crystal structure of the outer membrane decaheme cytochrome MtrC (A430Boc-Lys) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Nash, B.W, Lockwood, C.J, Whiting, K, Butt, J.N, Clarke, T.A, Edwards, M.J. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Genetic Code Expansion in Shewanella oneidensis MR-1 Allows Site-Specific Incorporation of Bioorthogonal Functional Groups into a c -Type Cytochrome.
Acs Synth Biol, 13, 2024
|
|
8QC9
 
 | | Crystal structure of the outer membrane decaheme cytochrome MtrC (A293Boc-Lys) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Nash, B.W, Lockwood, C.J, Edwards, M.J, Whiting, K, Butt, J.N, Clarke, T.A. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetic Code Expansion in Shewanella oneidensis MR-1 Allows Site-Specific Incorporation of Bioorthogonal Functional Groups into a c -Type Cytochrome.
Acs Synth Biol, 13, 2024
|
|
6O3J
 
 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
8S65
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) as target for anti Toxoplasma gondii compounds: crystal structure, biochemical characterization and biological evaluation of inhibitors | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Mazzone, F, Hoeppner, A, Reiners, J, Applegate, V, Abdullaziz, M, Gottstein, J, Wesemann, M, Kurz, T, Smits, S.H, Pfeffer, K. | | Deposit date: | 2024-02-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | SOLUTION SCATTERING (2.56 Å), X-RAY DIFFRACTION | | Cite: | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase as target for anti Toxoplasma gondii agents: crystal structure, biochemical characterization and biological evaluation of inhibitors.
Biochem.J., 481, 2024
|
|
7LNM
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
8RW3
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with a non-covalent 1,2- Cyclophellitol aziridine | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Moran, E, Davies, G, Ofamn, T, Heming, J, Nin-Hill, A, Kullmer, F, Steneker, R, Klein, A, Bennett, M, Ruijgrok, G, Kok, K, Aerts, J, Van der Marel, G, Rovira, C, Artola, M, Codee, J, Overkleeft, H. | | Deposit date: | 2024-02-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
5NPE
 
 | | Crystal Structure of cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with beta Cyclophellitol Aziridine probe KY358 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
6FSE
 
 | | Mus musculus acetylcholinesterase in complex with 1-(4-(4-Ethylpiperazin-1-yl)piperidin-1-yl)-2-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)ethanone dihydrochloride (15) | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]-2-[4-(4-methoxyphenyl)phenoxy]ethanone, 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
7KH6
 
 | |
