3MJI
 
 | |
4UHC
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (Native) | | Descriptor: | CHLORIDE ION, ESTERASE | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
3DAY
 
 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP-CPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-coenzyme A synthetase ACSM2A, mitochondrial precursor, ... | | Authors: | Pilka, E.S, Kochan, G.T, Yue, W.W, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
6ACJ
 
 | |
3MKF
 
 | | SHV-1 beta-lactamase complex with GB0301 | | Descriptor: | ({[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}methyl)boronic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Ke, W. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3MND
 
 | |
3DWJ
 
 | | Heme-proximal W188H mutant of inducible nitric oxide synthase | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, AMMONIUM ION, Nitric oxide synthase, ... | | Authors: | Tejero, J, Biswas, A, Wang, Z.-Q, Haque, M.M, Hemann, C, Zweier, J.L, Page, R.C, Misra, S, Stuehr, D.J. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Stabilization and Characterization of a Heme-Oxy Reaction Intermediate in Inducible Nitric-oxide Synthase
J.Biol.Chem., 283, 2008
|
|
4KRW
 
 | |
3MNJ
 
 | | Human Carbonic Anhydrase II Mutant K170E | | Descriptor: | Carbonic anhydrase 2, SODIUM ION, ZINC ION | | Authors: | Domsic, J.F, McKenna, R. | | Deposit date: | 2010-04-21 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and kinetic study of the extended active site for proton transfer in human carbonic anhydrase II.
Biochemistry, 49, 2010
|
|
3MNU
 
 | |
3DBP
 
 | | Crystal Structure of Human Orotidine 5'-Monophosphate Decarboxylase Complexed with 6-NH2-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Liu, Y, To, T, Bello, A.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-Monophosphate Decarboxylase Complexed with 6-NH2-UMP
To be Published
|
|
4KLW
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 2-(aminocarbonyl)benzoate | | Descriptor: | 1,2-ETHANEDIOL, 2-carbamoylbenzoic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
3DC3
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
3MKZ
 
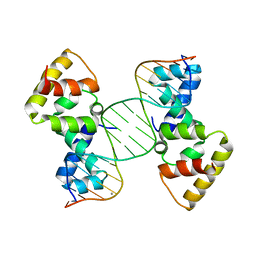 | | Structure of SopB(155-272)-18mer complex, P21 form | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3DXM
 
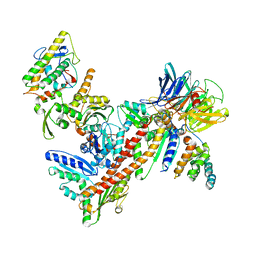 | | Structure of Bos taurus Arp2/3 Complex with Bound Inhibitor CK0993548 | | Descriptor: | (2S)-2-(3-bromophenyl)-3-(5-chloro-2-hydroxyphenyl)-1,3-thiazolidin-4-one, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Nolen, B.J, Tomasevic, N, Russell, A, Pierce, D.W, Jia, Z, Hartman, J, Sakowicz, R, Pollard, T.D. | | Deposit date: | 2008-07-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nature, 460, 2009
|
|
3ML8
 
 | |
3MO5
 
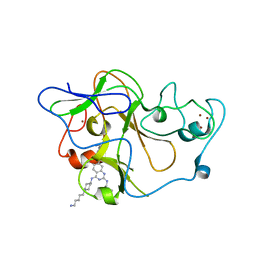 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E72 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-[1-(5-aminopentyl)piperidin-4-yl]-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
4KTJ
 
 | |
3DDR
 
 | |
4UNB
 
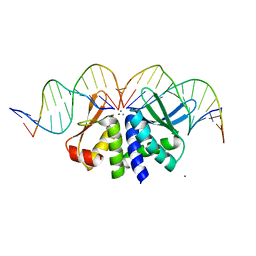 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 6 DAYS INCUBATION IN 5MM MN (STATE 5) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3DEF
 
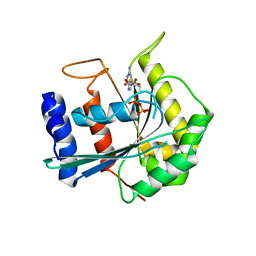 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
4KMP
 
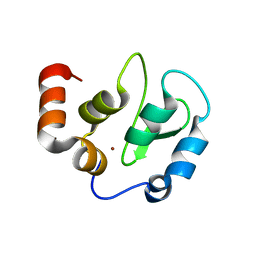 | | Structure of XIAP-BIR3 and inhibitor | | Descriptor: | (2S,2'S)-N,N'-[(6,6'-difluoro-1H,1'H-2,2'-biindole-3,3'-diyl)bis{methanediyl[(2R,4S)-4-hydroxypyrrolidine-2,1-diyl][(2S)-1-oxobutane-1,2-diyl]}]bis[2-(methylamino)propanamide], E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of XIAP-BIR3 and inhibitor
To be Published
|
|
3DYJ
 
 | |
3MLM
 
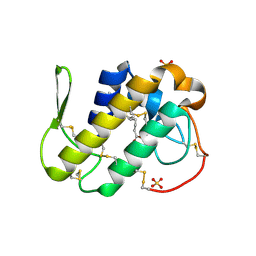 | | Crystal structure of Bn IV in complex with myristic acid: A Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom | | Descriptor: | BN-IV Lys-49 Phospholipase A2, MYRISTIC ACID, SULFATE ION | | Authors: | Delatorre, P, Rocha, B.A.M, Cavada, B.S, Toyama, M.H, Toyama, D, Gadelha, C.A.A. | | Deposit date: | 2010-04-17 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bn IV in complex with myristic acid: a Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom.
Biochimie, 93, 2011
|
|
4KN4
 
 | |
