6HKE
 
 | | MatC (Rpa3494) from Rhodopseudomonas palustris with bound malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, D-MALATE, Possible TctC subunit of the Tripartite Tricarboxylate Transport(TTT) Family | | Authors: | Rosa, L.T, Dix, S, Rafferty, J.B, Kelly, D.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A New Mechanism for High-Affinity Uptake of C4-Dicarboxylates in Bacteria Revealed by the Structure of Rhodopseudomonas palustris MatC (RPA3494), a Periplasmic Binding Protein of the Tripartite Tricarboxylate Transporter (TTT) Family.
J. Mol. Biol., 431, 2019
|
|
6CVA
 
 | |
6NNC
 
 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NMO
 
 | |
6R0H
 
 | | Glycogen Phosphorylase b in complex with 3 | | Descriptor: | 3-(4-fluorophenyl)-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]benzamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Tsagkarakou, S.A, Koulas, M.S, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
6R0I
 
 | | Glycogen Phosphorylase b in complex with 4 | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Koulas, M.S, Tsagkarakou, S.A, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
5EDR
 
 | | EGFR kinase (T790M/L858R) with inhibitor compound 27: ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Descriptor: | Epidermal growth factor receptor, ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6R9X
 
 | | Human Cyclophilin D in complex with N-cyclopentyl-N'-pyridin-2-ylmethyl-oxalamide | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ~{N}'-cyclopentyl-~{N}-(pyridin-2-ylmethyl)ethanediamide | | Authors: | Graedler, U. | | Deposit date: | 2019-04-04 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R8L
 
 | | Human Cyclophilin D in complex with 1-((1S,9S,10S)-10-Hydroxy-12-oxa-8-aza-tricyclo[7.3.1.02,7]trideca-2,4,6-trien-4-ylmethyl)-3- {2-[(R)-2-(2-methylsulfanyl-phenyl)-pyrrolidin-1-yl]-2-oxo-ethyl}-urea | | Descriptor: | 1-[2-[(2~{R})-2-(2-methylsulfanylphenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-3-[[(1~{S},9~{S},10~{S})-10-oxidanyl-12-oxa-8-azatricyclo[7.3.1.0^{2,7}]trideca-2(7),3,5-trien-4-yl]methyl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R8O
 
 | | Human Cyclophilin D in complex with 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea | | Descriptor: | 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R8W
 
 | | Human Cyclophilin D in complex with 2-(exo-3,5-Dioxo-4-aza-tricyclo[5.2.1.02,6]dec-4-yl)-N-((1R,9R,10S)-10-hydroxy-12-oxa-8-aza-tricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ylmethyl)-acetamide | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2-(exo-3,5-Dioxo-4-aza-tricyclo[5.2.1.02,6]dec-4-yl)-N-((1R,9R,10S)-10-hydroxy-12-oxa-8-aza-tricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ylmethyl)-acetamide, HEXAETHYLENE GLYCOL, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6R9U
 
 | | Human Cyclophilin D in complex with fragment | | Descriptor: | 14-ethyl-4,6-dioxa-10,14-diazatricyclo[7.6.0.0^{3,7}]pentadeca-1(9),2,7-trien-13-one, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-04 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8FA0
 
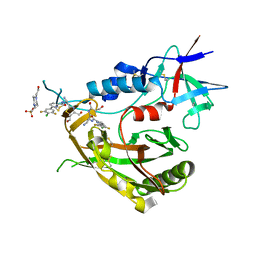 | |
1CP8
 
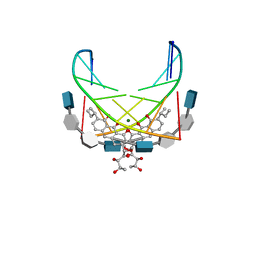 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
5F0A
 
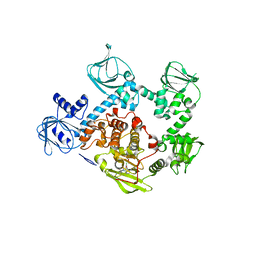 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | Authors: | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
8A9M
 
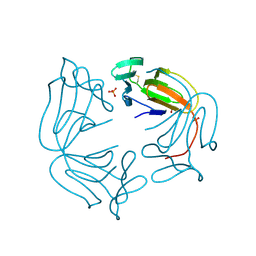 | | Hippeastrum hybrid agglutinin, HHA, complex with beta-mannose | | Descriptor: | Agglutinin, PHOSPHATE ION, beta-D-mannopyranose | | Authors: | Rizkallah, P.J. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure solution of amaryllis lectin by molecular replacement with only 4% of the total diffracting matter.
Acta Crystallogr D Biol Crystallogr, 52, 1996
|
|
3O4T
 
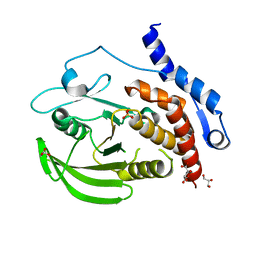 | |
6OJU
 
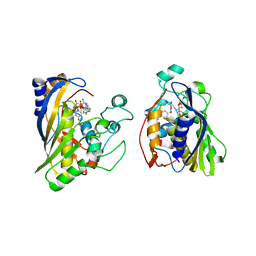 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
7QEV
 
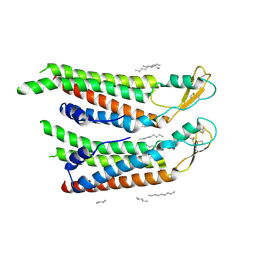 | | human Connexin 26 at 55mm Hg PCO2, pH7.4:two masked subunits, class D | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
5M44
 
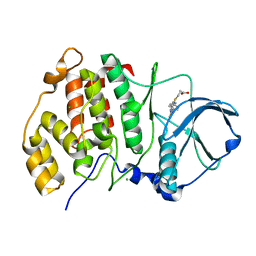 | | Complex structure of human protein kinase CK2 catalytic subunit with a thieno[2,3-d]pyrimidin inhibitor crystallized under high-salt conditions | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
8JHE
 
 | | Hyper-thermostable ancestral L-amino acid oxidase 2 (HTAncLAAO2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hyper thermostable ancestral L-amino acid oxidase | | Authors: | Kawamura, Y, Ishida, C, Miyata, R, Miyata, A, Hayashi, S, Fujinami, D, Ito, S, Nakano, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of hyper-thermostable ancestral L-amino acid oxidase that can convert Trp derivatives to D-forms by chemoenzymatic reaction.
Commun Chem, 6, 2023
|
|
6ZJQ
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6ZJW
 
 | |
5M4C
 
 | | Complex structure of human protein kinase CK2 catalytic subunit with a thieno[2,3-d]pyrimidin inhibitor crystallized under low-salt conditions | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
6OJS
 
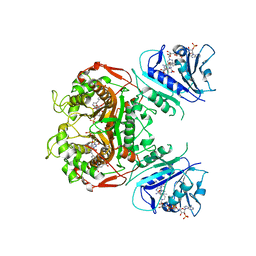 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP, MTX and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.J, Anderson, K.S, Jorgensen, W.L, Valhondo, M. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
