3EM6
 
 | | Crystal structure of I50L/A71V mutant of hiv-1 protease in complex with inhibitor darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Royer, C.J, King, N.M, Prabu-Jeyabalan, M, Ng, C, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies on atazanavir specific I50L drug-resistant HIV-1 protease mutant.
To be Published
|
|
3EMM
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 with Bound Heme | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Uncharacterized protein At1g79260 | | Authors: | Bianchetti, C.M, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
3E3D
 
 | | Structure of hen egg white lysozyme with the magic triangle I3C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Lysozyme C | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-07 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E3P
 
 | |
3E41
 
 | | Q138F HincII bound to GTCGAC and 5 mM Ca2+ | | Descriptor: | 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Horton, N.C, Babic, A.C, Little, E.J, Manohar, V.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | DNA distortion and specificity in a sequence-specific endonuclease.
J.Mol.Biol., 383, 2008
|
|
3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
3EES
 
 | |
3E60
 
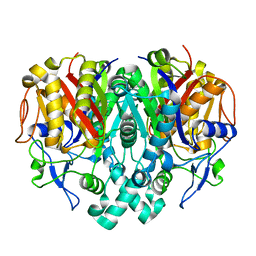 | |
3E7B
 
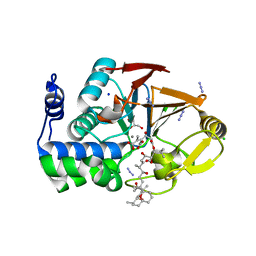 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10R)-10-{(2S,3S,6R,8S,9R)-3,9-dimethyl-8-[(3S)-3-methyl-4-oxopentyl]-1,7-dioxaspiro[5.5]undec-2-yl}-3,7-dihydroxy-2-methoxy-6-methyl-1-(1-methylethyl)-5-oxoundecyl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
3EGE
 
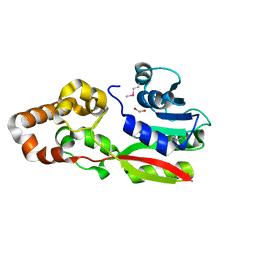 | |
3E7L
 
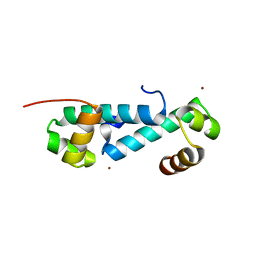 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | Descriptor: | Transcriptional regulator (NtrC family), ZINC ION | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
3EGQ
 
 | |
3E87
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta peptide, N-[(1S)-2-amino-1-phenylethyl]-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)thiophene-2-carboxamide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8X
 
 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
3EJF
 
 | |
3EJL
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, PHOSPHATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-09-18 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3EL2
 
 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EBF
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | (3R)-3-[(1,2,3,4-tetrahydroisoquinolin-7-yloxy)methyl]-2,3-dihydrothieno[2,3-f][1,4]oxazepin-5-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3ENK
 
 | |
3EBV
 
 | | Crystal structure of putative Chitinase A from Streptomyces coelicolor. | | Descriptor: | Chinitase A, SULFATE ION | | Authors: | Vigdorovich, V, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative Chitinase A from Streptomyces coelicolor
To be Published
|
|
3EOX
 
 | |
3EP9
 
 | | Human AdoMetDC with no putrescine bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
3EPM
 
 | | Crystal structure of Caulobacter crescentus ThiC | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SULFATE ION, Thiamine biosynthesis protein thiC, ... | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3EQ2
 
 | |
