8CK8
 
 | |
8CK4
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (4S)-1-(3,5-difluorophenyl)-5,5-difluoro-3-methanesulfonyl-4,5,6,7-tetrahydro-2-benzothiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-6,7-dihydro-4~{H}-2-benzothiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Musil, D. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
4YFF
 
 | | TNNI3K complexed with inhibitor 2 | | Descriptor: | 3-[(5-bromo-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
6J8Q
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAI
 
 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
6JN3
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor MS05 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Serine Beta-Lactamase KPC-2, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6KL6
 
 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
7KQD
 
 | | Prefusion RSV F Bound to RV521 | | Descriptor: | 1'-{[5-(aminomethyl)-1-(4,4,4-trifluorobutyl)-1H-benzimidazol-2-yl]methyl}-6'-fluorospiro[cyclopropane-1,3'-indol]-2'(1'H)-one, Fusion glycoprotein F0, SULFATE ION | | Authors: | McLellan, J.S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Discovery of Sisunatovir (RV521), an Inhibitor of Respiratory Syncytial Virus Fusion.
J.Med.Chem., 64, 2021
|
|
7MGJ
 
 | |
7MGK
 
 | |
7LGK
 
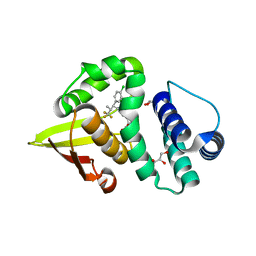 | | Crystal structure of soluble guanylate cyclase activator runcaciguat (BAY 1101042) bound to nostoc H-NOX domain | | Descriptor: | CHLORIDE ION, GLYCEROL, H-NOX domain protein, ... | | Authors: | van den Akker, F, Kumar, V, Schaefer, M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the Soluble Guanylate Cyclase Activator Runcaciguat (BAY 1101042).
J.Med.Chem., 64, 2021
|
|
6V79
 
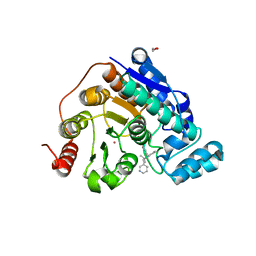 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03951526 Å) | | Cite: | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
6KL5
 
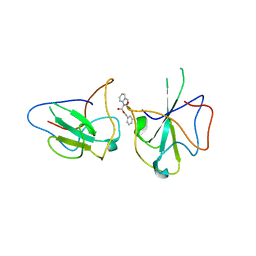 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
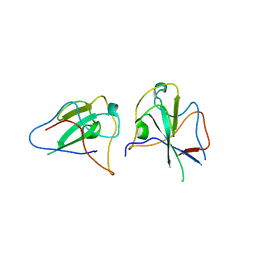 | |
7Z59
 
 | | SARS-CoV-2 main protease (Mpro) covalently modified with a penicillin derivative | | Descriptor: | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Owen, C.D, Malla, T.R, Brewitz, L, Lukacik, P, Strain-Damerell, C, Mikolajek, H, Muntean, D.G, Aslam, H, Salah, E, Tumber, A, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
J.Med.Chem., 65, 2022
|
|
7ZAA
 
 | | BRD4 in complex with FragLite11 | | Descriptor: | GLYCEROL, Isoform C of Bromodomain-containing protein 4, ~{N}-(4-bromophenyl)ethanamide | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAJ
 
 | | BRD4 in complex with FragLite16 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZA6
 
 | | BRD4 in complex with FragLite4 | | Descriptor: | 4-bromanylpyridin-2-amine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAT
 
 | | BRD4 in complex with FragLite20 | | Descriptor: | 4-bromanyl-2-(methoxymethyl)pyridine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAD
 
 | | BRD4 in complex with FragLite12 | | Descriptor: | 4-iodanylbenzamide, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAE
 
 | | BRD4 in complex with FragLite15 | | Descriptor: | 4-bromanyl-2-methoxy-pyridine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAR
 
 | | BRD4 in complex with FragLite18 | | Descriptor: | (4-bromanylpyridin-2-yl)methanol, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
