3ITJ
 
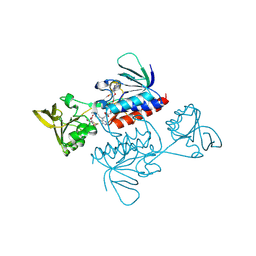 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | Authors: | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
5BKN
 
 | |
5TMF
 
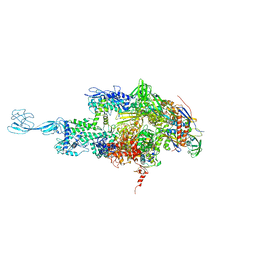 | | Re-refinement of thermus thermophilus RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
5AN1
 
 | |
1PN0
 
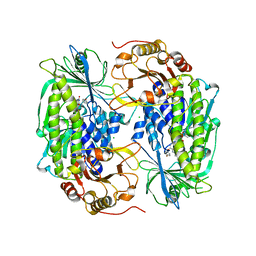 | | Phenol hydroxylase from Trichosporon cutaneum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, ... | | Authors: | Enroth, C. | | Deposit date: | 2003-06-12 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of phenol hydroxylase and correction of sequence errors.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2L7L
 
 | |
1R51
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 8-AZAXANTHIN | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Prange, T, Retailleau, P, Colloc'h, N. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5OFT
 
 | |
4DPE
 
 | | Structure of MMP3 complexed with a platinum-based inhibitor. | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Belviso, B.D, Arnesano, F, Calderone, V, Caliandro, R, Natile, G, Siliqi, D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of matrix metalloproteinase-3 with a platinum-based inhibitor.
Chem.Commun.(Camb.), 49, 2013
|
|
1H33
 
 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
1H32
 
 | | Reduced SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME C, DIHEME CYTOCHROME C, ... | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H31
 
 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
3IFK
 
 | |
4FGW
 
 | | Structure of Glycerol-3-Phosphate Dehydrogenase, GPD1, from Sacharomyces Cerevisiae | | Descriptor: | Glycerol-3-phosphate dehydrogenase [NAD(+)] 1 | | Authors: | Aparicio, D, Munmun, N, Carpena, X, Fita, I, Loewen, P. | | Deposit date: | 2012-06-05 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase (GPD1) from Saccharomyces cerevisiae at 2.45A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7E0M
 
 | | Crystal structure of phospholipase D | | Descriptor: | Phospholipase, SULFATE ION | | Authors: | Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket.
Int J Mol Sci, 22, 2021
|
|
3RN8
 
 | |
4G9L
 
 | | Structure of MMP3 complexed with NNGH inhibitor. | | Descriptor: | CALCIUM ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, Stromelysin-1, ... | | Authors: | Belviso, B.D, Arnesano, F, Calderone, V, Caliandro, R, Natile, G, Siliqi, D. | | Deposit date: | 2012-07-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of matrix metalloproteinase-3 with a platinum-based inhibitor.
Chem.Commun.(Camb.), 49, 2013
|
|
6KTK
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
3RNN
 
 | |
6KTJ
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
4TYD
 
 | | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease | | Descriptor: | (4R,6S,7Z,15S,17S)-17-[({7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)methyl]-13-methyl-N-[(1-methylcyclopropyl)sulfonyl]-2,14-dioxo-1,3,13-triazatricyclo[13.2.0.0~4,6~]heptadec-7-ene-4-carboxamide, CHLORIDE ION, NS3 protease, ... | | Authors: | Parsy, C. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4HNN
 
 | |
2OQY
 
 | | The crystal structure of muconate cycloisomerase from Oceanobacillus iheyensis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Toro, R, Fedorov, E.V, Bonanno, J, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
