3SRP
 
 | |
1G42
 
 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | Descriptor: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
4LD1
 
 | |
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PHG
 
 | | Crystal structure of Ypt7 covalently modified with GTP | | Descriptor: | GTP-binding protein YPT7, MAGNESIUM ION, N-[3-(propanoylamino)propyl]guanosine 5'-(tetrahydrogen triphosphate), ... | | Authors: | Koch, D, Wiegandt, D, Vieweg, S, Hofmann, F, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
4EDV
 
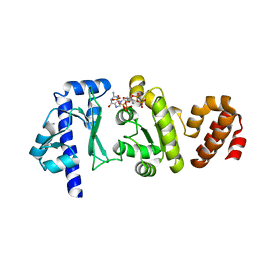 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to pppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EE7
 
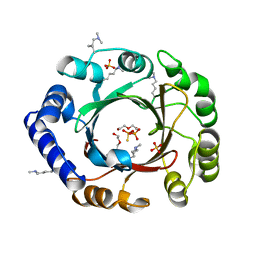 | |
1C8R
 
 | | BACTERIORHODOPSIN D96N BR STATE AT 2.0 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, PROTEIN (BACTERIORHODOPSIN), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
2JIP
 
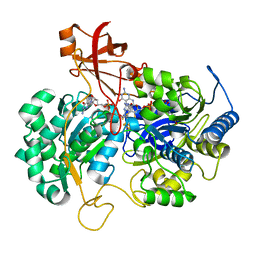 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIO
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIQ
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
1KQF
 
 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARDIOLIPIN, FORMATE DEHYDROGENASE, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5T55
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH GLOBOTETRAOSE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
1AA6
 
 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
2C6H
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
4QXX
 
 | |
4N7U
 
 | |
3SAO
 
 | | The Siderocalin Ex-FABP functions through dual ligand specificities | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2,3-DIHYDROXY-BENZOIC ACID, Extracellular fatty acid-binding protein, ... | | Authors: | Correnti, C, Strong, R.K, Clifton, M.C. | | Deposit date: | 2011-06-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Galline Ex-FABP Is an Antibacterial Siderocalin and a Lysophosphatidic Acid Sensor Functioning through Dual Ligand Specificities.
Structure, 19, 2011
|
|
3SWS
 
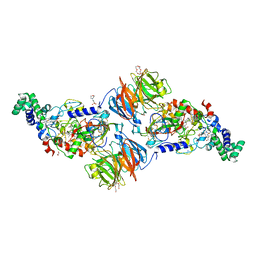 | |
4C3L
 
 | | Structure of wildtype PII from S. elongatus at high resolution | | Descriptor: | CHLORIDE ION, NITROGEN REGULATORY PROTEIN P-II, TETRAETHYLENE GLYCOL | | Authors: | Zeth, K, Forchhammer, K. | | Deposit date: | 2013-08-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis and Target-Specific Modulation of Adp Sensing by the Synechococcus Elongatus Pii Signaling Protein.
J.Biol.Chem., 289, 2014
|
|
4BOF
 
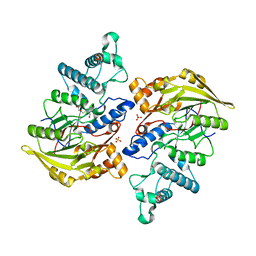 | | Crystal structure of arginine deiminase from group A streptococcus | | Descriptor: | ARGININE DEIMINASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Henningham, A, Ericsson, D.J, Langer, K, Casey, L, Jovcevski, B, Chhatwal, G.S, Aquilina, J.A, Batzloff, M.R, Kobe, B, Walker, M.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus.
MBio, 4, 2013
|
|
5T36
 
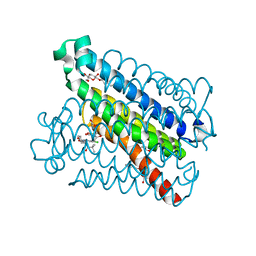 | | Crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 4-chloro-2-[({(1S,2S)-2-[(2,2-dimethylpropanoyl)amino]cyclopentyl}methyl)amino]benzoic acid, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2E9U
 
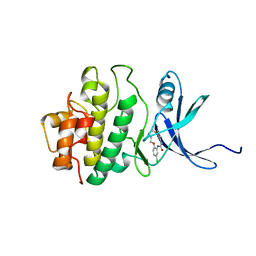 | | Structure of h-CHK1 complexed with A780125 | | Descriptor: | 18-CHLORO-11,12,13,14-TETRAHYDRO-1H,10H-8,4-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECIN-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2JIM
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
