4HW6
 
 | |
4HPE
 
 | |
4G5A
 
 | |
4FSV
 
 | |
4FUU
 
 | |
4FTD
 
 | |
4GHB
 
 | |
4FS7
 
 | |
4G2A
 
 | |
4IB2
 
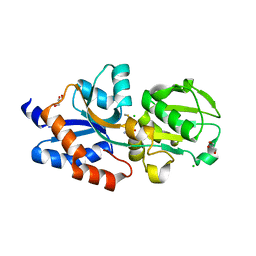 | |
4IAJ
 
 | |
4HSP
 
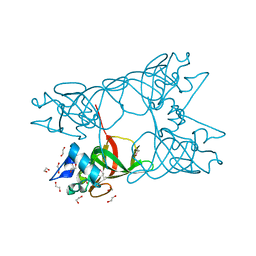 | |
3F7P
 
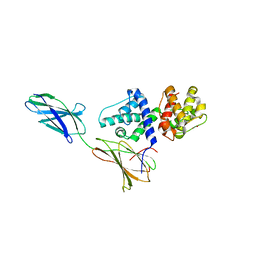 | |
3FDD
 
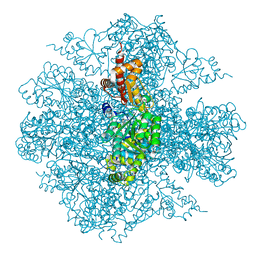 | | The Crystal Structure of the Pseudomonas dacunhae Aspartate-Beta-Decarboxylase Reveals a Novel Oligomeric Assembly for a Pyridoxal-5-Phosphate Dependent Enzyme | | Descriptor: | ACETATE ION, CHLORIDE ION, L-aspartate-beta-decarboxylase, ... | | Authors: | Lima, S, Sundararaju, B, Huang, C, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the Pseudomonas dacunhae aspartate-beta-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme.
J.Mol.Biol., 388, 2009
|
|
3FM8
 
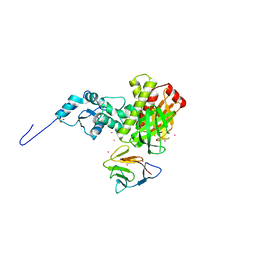 | | Crystal structure of full length centaurin alpha-1 bound with the FHA domain of KIF13B (CAPRI target) | | Descriptor: | Centaurin-alpha-1, Kinesin-like protein KIF13B, SULFATE ION, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3FO4
 
 | |
3EZG
 
 | |
3GOT
 
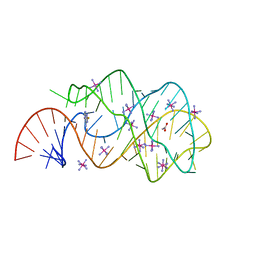 | | Guanine riboswitch C74U mutant bound to 2-fluoroadenine. | | Descriptor: | 2-fluoroadenine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3H3K
 
 | |
3H90
 
 | |
3HDK
 
 | | Crystal structure of chemically synthesized [Aib51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [Aib51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-28 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HD7
 
 | | HELICAL EXTENSION OF THE NEURONAL SNARE COMPLEX INTO THE MEMBRANE, spacegroup C 1 2 1 | | Descriptor: | SULFATE ION, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3G5U
 
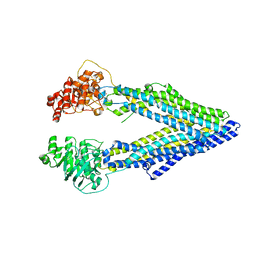 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3GER
 
 | | Guanine riboswitch bound to 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Batey, R.T. | | Deposit date: | 2009-02-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3G0B
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with TAK-322 | | Descriptor: | 2-({6-[(3R)-3-aminopiperidin-1-yl]-3-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl}methyl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
