1YWC
 
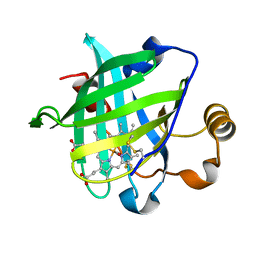 | | Structure of the ferrous CO complex of NP4 from Rhodnius Prolixus at pH 7.0 | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, nitrophorin 4 | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
3RWN
 
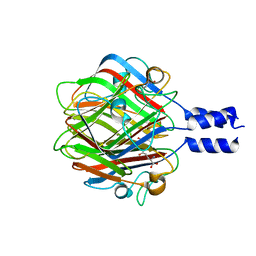 | |
1KGK
 
 | | Direct Observation of a Cytosine Analog that Forms Five Hydrogen Bonds to Guanosine; Guanyl G-Clamp | | Descriptor: | 5'-D(*GP*(GCK)P*GP*TP*AP*TP*AP*CP*GP*C)-3', METHOXY-ETHOXYL, SPERMINE (FULLY PROTONATED FORM) | | Authors: | Wilds, C.J, Maier, M.A, Tereshko, V, Manoharan, M, Egli, M. | | Deposit date: | 2001-11-27 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct Observation of a Cytosine Analogue that Forms Five Hydrogen Bonds to Guanosine: Guanidino G-Clamp
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
6IXD
 
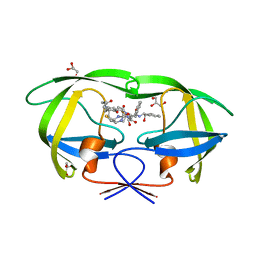 | | X-ray crystal structure of bPI-11 hiv-1 protease complex | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adachi, M, Hidaka, K. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Acquired Removability of Aspartic Protease Inhibitors by Direct Biotinylation.
Bioconjug.Chem., 30, 2019
|
|
4XZH
 
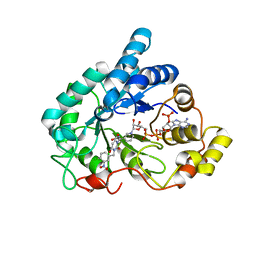 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0048 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-chloro-3-nitrobenzyl)-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
3DVZ
 
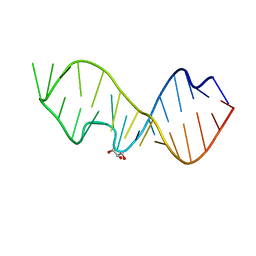 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3JYO
 
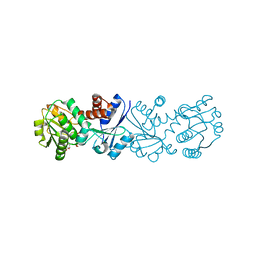 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Niefind, K, Schomburg, D. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
5SAO
 
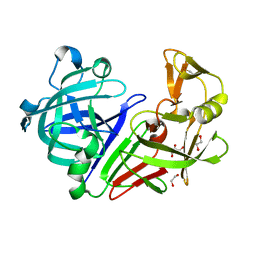 | | Endothiapepsin in complex with compound FU58-1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8R)-2-({[(3,5-dimethyl-1,2-oxazol-4-yl)methyl](methyl)amino}methyl)-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]pyrimidin-4-amine, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAL
 
 | | Endothiapepsin in complex with compound FU5-2 | | Descriptor: | (1Z)-1-imino-1H-isoindol-3-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7UYD
 
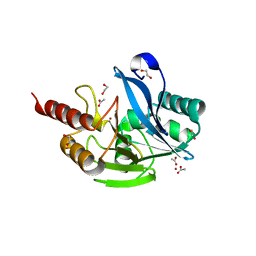 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7T89
 
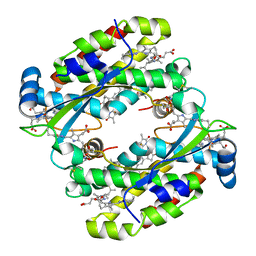 | | Light harvesting complex Phycocyanin PC577 from the cryptophyte Hemiselmis pacifica CCMP 706 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Michie, K.A, Curmi, P.C, Harrop, S, Rathbone, H.W. | | Deposit date: | 2021-12-16 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
6MZ1
 
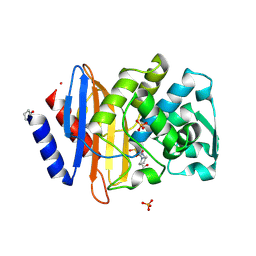 | |
5M2K
 
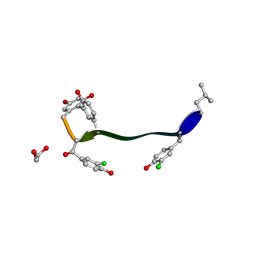 | | Crystal structure of vancomycin-Zn(II) complex | | Descriptor: | 1,2-ETHANEDIOL, ZINC ION, vancomycin, ... | | Authors: | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
2VHA
 
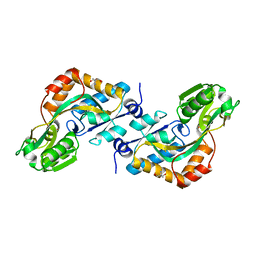 | | DEBP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | Authors: | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | Deposit date: | 2007-11-20 | | Release date: | 2008-07-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
1K4P
 
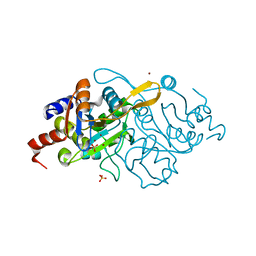 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with zinc ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, SULFATE ION, ZINC ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
6RD2
 
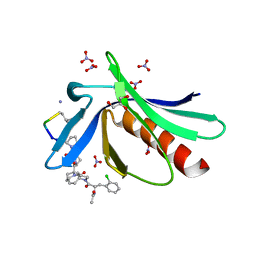 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1ZK4
 
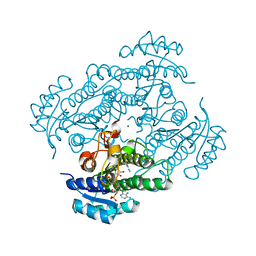 | | Structure of R-specific alcohol dehydrogenase (wildtype) from Lactobacillus brevis in complex with acetophenone and NADP | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
3DW6
 
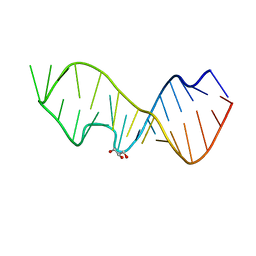 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
6IFP
 
 | |
1C0Q
 
 | | COMPLEX OF VANCOMYCIN WITH 2-ACETOXY-D-PROPANOIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin binding to low-affinity ligands: delineating a minimum set of interactions necessary for high-affinity binding.
J.Med.Chem., 42, 1999
|
|
3AUB
 
 | | A simplified BPTI variant stabilized by the A14G and A38V substitutions | | Descriptor: | Bovine Pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
4EA8
 
 | | X-ray crystal structure of PerB from Caulobacter crescentus in complex with coenzyme A and GDP-N-acetylperosamine at 1 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-N-acetylperosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
6FVI
 
 | |
6YF3
 
 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6PWS
 
 | |
