7LUX
 
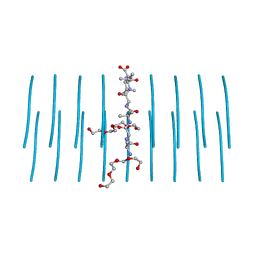 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
3L0F
 
 | |
5G3X
 
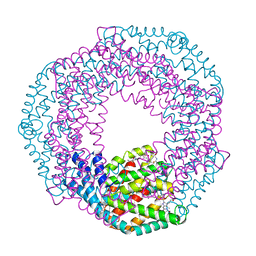
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7M02
 
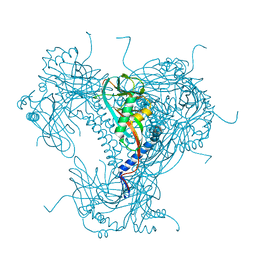
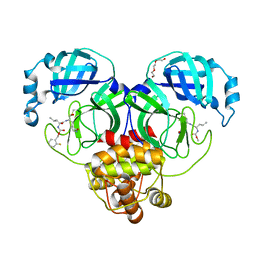 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 17c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
6G20
 
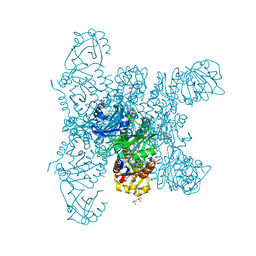 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its functional Meta-F intermediate state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Schmidt, A, Sauthof, L, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
7LZV
 
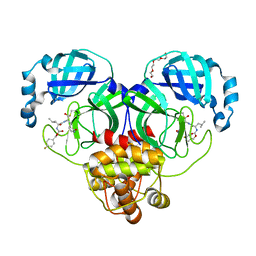 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 19b | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
6G3O
 
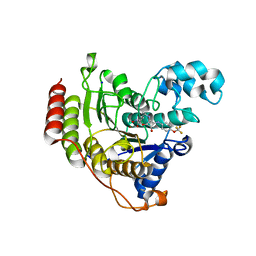 | | Crystal structure of human HDAC2 in complex with (R)-6-[3,4-Dioxo-2-(4-trifluoromethoxy-phenylamino)-cyclobut-1-enylamino]-heptanoic acid hydroxyamide | | Descriptor: | (6~{R})-6-[[3,4-bis(oxidanylidene)-2-[[4-(trifluoromethyloxy)phenyl]amino]cyclobuten-1-yl]amino]-~{N}-oxidanyl-heptanamide, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Isabet, T, Aurelly, M, Chantalat, L, Thoreau, E. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1HF4
 
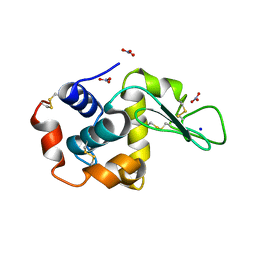 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8B21
 
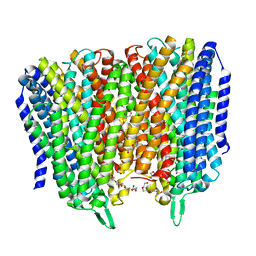 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
5DQF
 
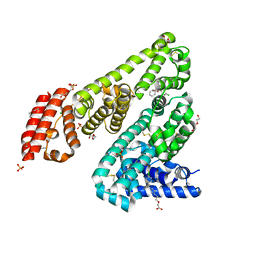 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
5HCG
 
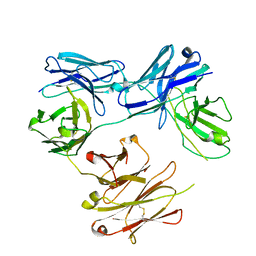 | | CRYSTAL STRUCTURE OF FREE-CODV. | | Descriptor: | CODV heavy-chain, CODV light chain | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
3OUJ
 
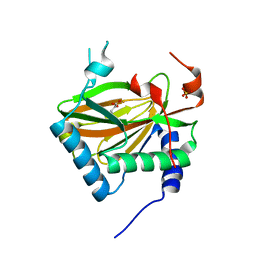 | | PHD2 with 2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Staker, B.L, Arakaki, T.L. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
6M9U
 
 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
6LM1
 
 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | Descriptor: | DECANE, DODECANE, HEXADECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
7LZX
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 1c | | Descriptor: | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
3OX7
 
 | | The crystal structure of uPA complex with peptide inhibitor MH027 at pH4.6 | | Descriptor: | MH027, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Andreasen, P.A, Huang, M.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The binding mechanism of a peptidic cyclic serine protease inhibitor
J.Mol.Biol., 412, 2011
|
|
7LZU
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 12b | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M04
 
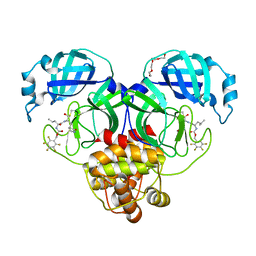 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 21c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
6GAQ
 
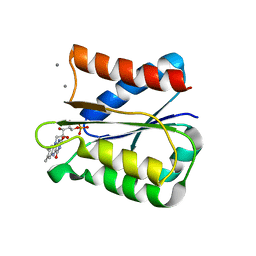 | | Crystal structure of oxidised Flavodoxin 2 from Bacillus cereus | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin, ... | | Authors: | Lofstad, M, Gudim, I, Hersleth, H.-P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Characterization of Different Flavodoxin Reductase-Flavodoxin (FNR-Fld) Interactions Reveals an Efficient FNR-Fld Redox Pair and Identifies a Novel FNR Subclass.
Biochemistry, 57, 2018
|
|
6MCQ
 
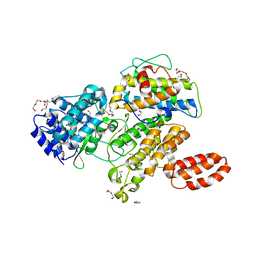 | | L. pneumophila effector kinase LegK7 in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2BHW
 
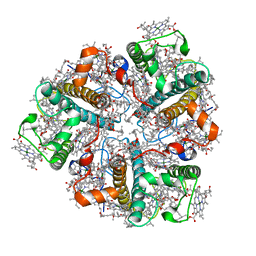 | | PEA LIGHT-HARVESTING COMPLEX II AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6'S,9R,9'R,13R,13'S)-4',5'-DIDEHYDRO-5',6',7',8',9,9',10,10',11,11',12,12',13,13',14,14',15,15'-OCTADECAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Standfuss, J, Terwisscha van Scheltinga, A.C, Lamborghini, M, Kuehlbrandt, W. | | Deposit date: | 2005-01-19 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of Photoprotection and Nonphotochemical Quenching in Pea Light-Harvesting Complex at 2.5 A Resolution.
Embo J., 24, 2005
|
|
7XJC
 
 | | Crystal structure of bacteriorhodopsin in the ground and K states after green laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
5HCO
 
 | | Endothiapepsin in complex with hydrazide | | Descriptor: | 4-(dimethylamino)benzohydrazide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
3N5C
 
 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
