8HMC
 
 | | base module state 1 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, Tetratricopeptide repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
4KER
 
 | | Crystal structure of SsoPox W263V | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
4KEU
 
 | | Crystal structure of SsoPox W263M | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
2YBG
 
 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YN1
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Gamma- Proteobacteria Common Ancestor (LGPCA) from the Precambrian Period | | Descriptor: | LGPCA THIOREDOXIN, TRIETHYLENE GLYCOL | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
7CGV
 
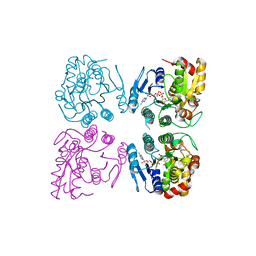 | | Full consensus L-threonine 3-dehydrogenase, FcTDH-IIYM (NAD+ bound form) | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Hiramatsu, N, Asano, Y, Nakano, S, Ito, S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Sequence Selection Method That Enables Full Consensus Design of Artificial l-Threonine 3-Dehydrogenases with Unique Enzymatic Properties.
Biochemistry, 59, 2020
|
|
7C7H
 
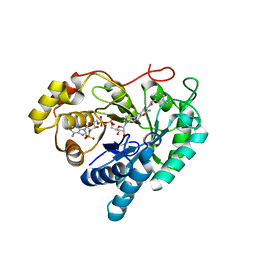 | |
4UZY
 
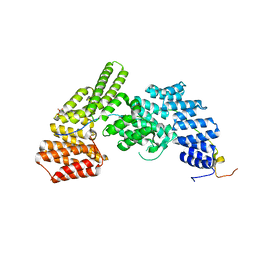 | | Crystal structure of the Chlamydomonas IFT70 and IFT52 complex | | Descriptor: | CITRATE ANION, FLAGELLAR ASSOCIATED PROTEIN, INTRAFLAGELLAR TRANSPORT PROTEIN IFT52, ... | | Authors: | Taschner, M, Lorentzen, E. | | Deposit date: | 2014-09-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Crystal Structures of Ift70/52 and Ift52/46 Provide Insight Into Intraflagellar Transport B Core Complex Assembly.
J.Cell Biol., 207, 2014
|
|
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
7C5Y
 
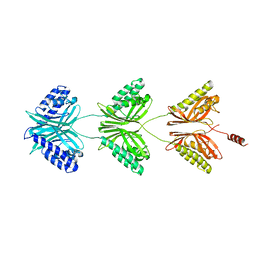 | |
7C5X
 
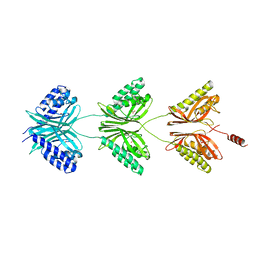 | |
6ESF
 
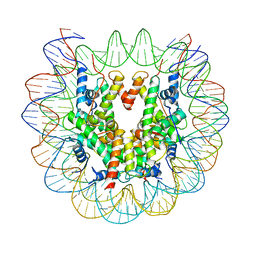 | | Nucleosome : Class 1 | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7QBN
 
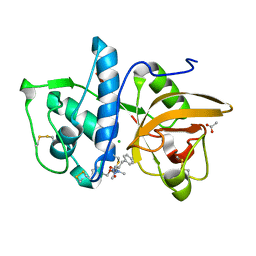 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBL
 
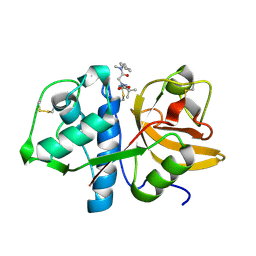 | |
7QBM
 
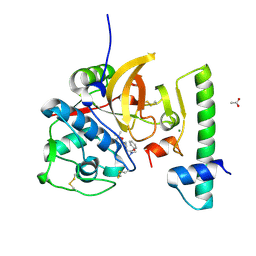 | | Structure of the activation intermediate of cathepsin K in complex with the 3-cyano-3-aza-beta-amino acid inhibitor Gu2602 | | Descriptor: | ACETATE ION, Cathepsin K, MAGNESIUM ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBO
 
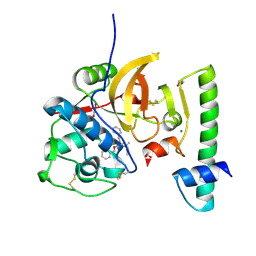 | | Structure of the activation intermediate of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
6ESG
 
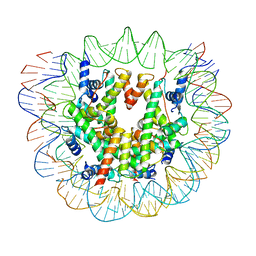 | | Nucleosome breathing : Class 2 | | Descriptor: | DNA (141-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6UVJ
 
 | |
6ESH
 
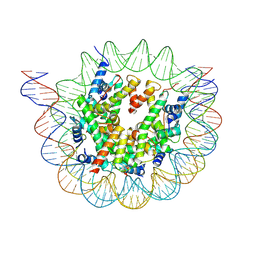 | | Nucleosome breathing : Class 3 | | Descriptor: | DNA (137-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7OHS
 
 | |
6ESI
 
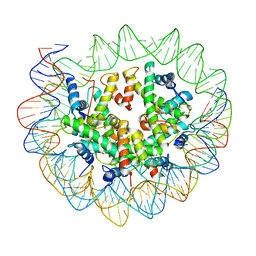 | | Nucleosome breathing : Class 4 | | Descriptor: | DNA (133-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6UWX
 
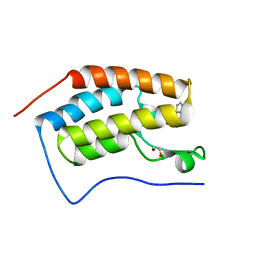 | |
4KZZ
 
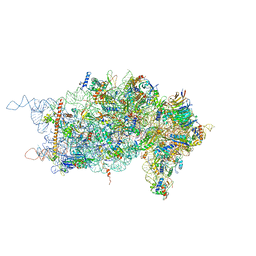 | |
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
8RU2
 
 | | Structure of the F-actin barbed end bound by formin mDia1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
