2N7J
 
 | |
2DWP
 
 | | A pseudo substrate complex of 6-phosphofructo-2-kinase of PFKFB | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, MAGNESIUM ION, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
1EQM
 
 | | CRYSTAL STRUCTURE OF BINARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE WITH ADENOSINE-5'-DIPHOSPHATE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiao, B, Blaszczyk, J, Ji, X. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR.
J.Biol.Chem., 276, 2001
|
|
2JM4
 
 | |
2E1Z
 
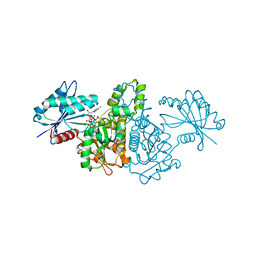 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with diadenosine tetraphosphate (Ap4A) obtained after co-crystallization with ATP | | Descriptor: | 1,2-ETHANEDIOL, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Propionate Kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-11-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Salmonella typhimurium propionate kinase and its complex with Ap4A: evidence for a novel Ap4A synthetic activity.
Proteins, 70, 2008
|
|
2PPB
 
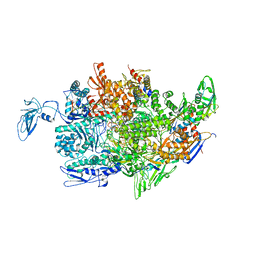 | | Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I, Landick, R. | | Deposit date: | 2007-04-28 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate loading in bacterial RNA polymerase.
Nature, 448, 2007
|
|
2FLY
 
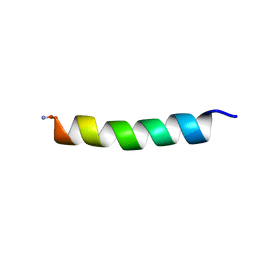 | | Proadrenomedullin N-Terminal 20 Peptide | | Descriptor: | Proadrenomedullin N-20 terminal peptide | | Authors: | Lucyk, S, Taha, H, Yamamoto, H, Miskolzie, M, Kotovych, G. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR conformational analysis of proadrenomedullin N-terminal 20 peptide, a proangiogenic factor involved in tumor growth
Biopolymers, 81, 2006
|
|
9C49
 
 | |
6ZQU
 
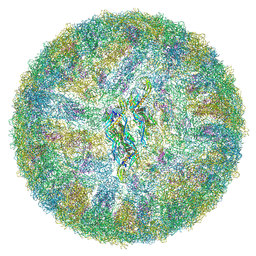 | | Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6I53
 
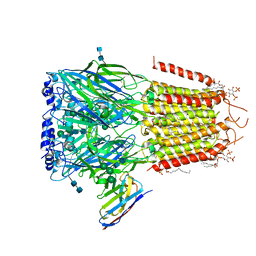 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
9FYP
 
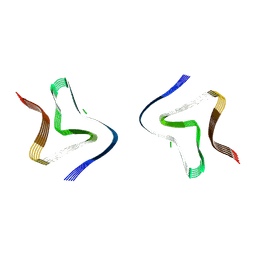 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
7AE1
 
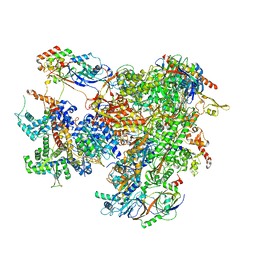 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9CJL
 
 | | Molecular basis of TMED9 dodecamer | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
9F6J
 
 | |
9F6L
 
 | |
9F6K
 
 | |
9BJO
 
 | | Cryo-EM of Azo-ffsy fiber | | Descriptor: | D-peptide ffsy | | Authors: | Zia, A, Guo, J, Xu, B, Wang, F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
9F6I
 
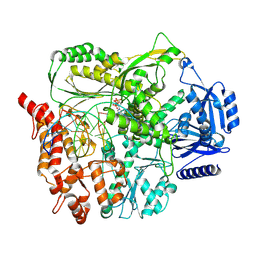 | |
9BJN
 
 | | Cryo-EM of Azo-ffspy fiber | | Descriptor: | D-peptide ffspy | | Authors: | Zia, A, Guo, J, Xu, B, Wang, F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
9F6E
 
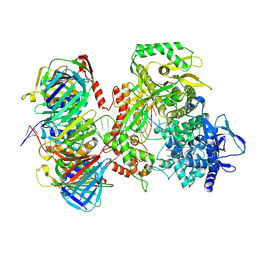 | | Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9F6D
 
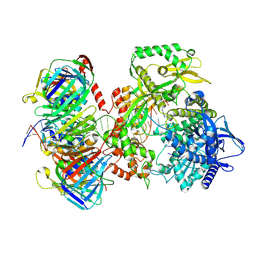 | | Human DNA polymerase epsilon bound to DNA and PCNA (open conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9F6F
 
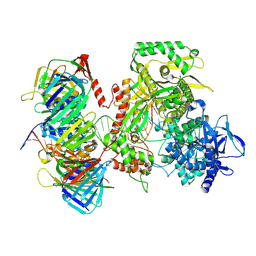 | | Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9B8S
 
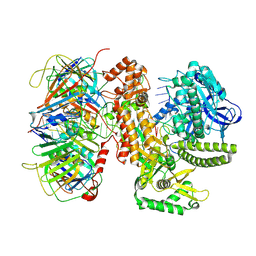 | | Human polymerase epsilon bound to PCNA and DNA in the nucleotide exchange state | | Descriptor: | DNA (5'-D(P*AP*AP*AP*GP*TP*GP*AP*AP*AP*AP*AP*TP*CP*TP*AP*AP*AP*GP*CP*AP*TP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*AP*TP*GP*CP*TP*TP*TP*AP*GP*AP*TP*TP*TP*TP*TP*C)-3'), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2024-03-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Structures of the human leading strand Pol epsilon-PCNA holoenzyme.
Nat Commun, 15, 2024
|
|
7A25
 
 | |
