4J3N
 
 | | Human Topoisomerase Iibeta in complex with DNA | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2013-02-06 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
7NB7
 
 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
3WBA
 
 | |
7NIE
 
 | |
5TMB
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
3WC4
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
7AIG
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-GLUTAMATE TENOFOVIR | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
4IRP
 
 | | Crystal structure of catalytic domain of human beta1,4-galactosyltransferase-7 in open conformation with manganses and UDP | | Descriptor: | ACETATE ION, Beta-1,4-galactosyltransferase 7, IMIDAZOLE, ... | | Authors: | Tsutsui, Y, Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2013-01-15 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-1,4-Galactosyltransferase 7 Enzyme Reveal Conformational Changes and Substrate Binding.
J.Biol.Chem., 288, 2013
|
|
6PN0
 
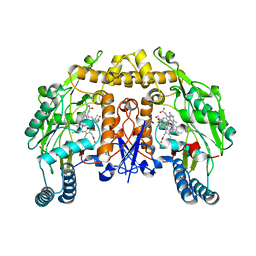 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6PNA
 
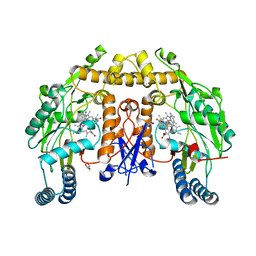 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(4-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[4-(2-aminoethyl)phenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
3OQ3
 
 | |
3WMZ
 
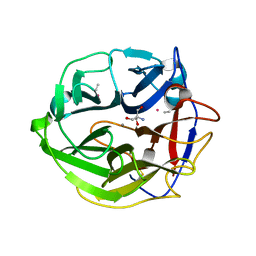 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase ethylmercury derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
6P1I
 
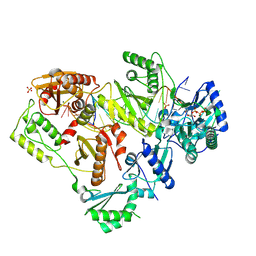 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
4IRW
 
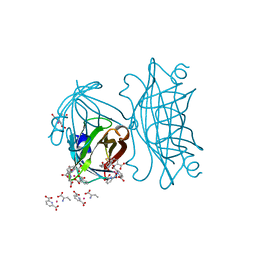 | | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form | | Descriptor: | BIOTIN, PYRIDINE-2,6-DICARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Bandara, R.A.M.S.S, Liu, D.Q, Hindupur, A, Tesh, K.F, Fox, R.O. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form
To be Published
|
|
5EUL
 
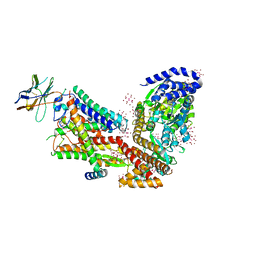 | | Structure of the SecA-SecY complex with a translocating polypeptide substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AYC08, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Li, L, Park, E, Ling, J, Ingram, J, Ploegh, H, Rapoport, T.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of a substrate-engaged SecY protein-translocation channel.
Nature, 531, 2016
|
|
7AJB
 
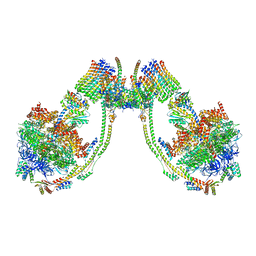 | | bovine ATP synthase dimer state1:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5TB4
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 4) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5CH4
 
 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
5T5L
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5C1S
 
 | |
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QSN
 
 | |
3WFA
 
 | | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase | | Descriptor: | Alpha-glucosidase, SODIUM ION, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Okuyama, M, Yoshida, T, Hondoh, H, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase
To be Published
|
|
8QSM
 
 | |
