7VL5
 
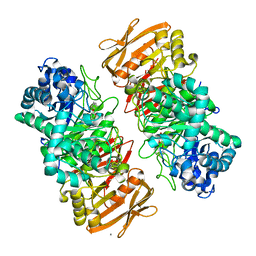 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with n-octyl-beta-D-glucoside | | Descriptor: | Beta-galactosidase, CALCIUM ION, octyl beta-D-glucopyranoside | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7S90
 
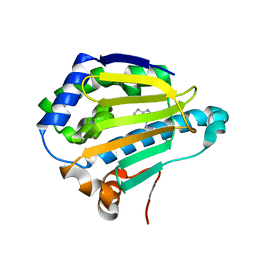 | | Cryogenic Human Hsp90a-NTD bound to adenine | | Descriptor: | ADENINE, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Lopez, K, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S8Y
 
 | |
6UO2
 
 | |
8P8S
 
 | | Recombinant Ym2 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 4 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, A, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
7VL7
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with esculin | | Descriptor: | 6-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-7-oxidanyl-chromen-2-one, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
6TW0
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6UKU
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 3 | | Descriptor: | 4,4'-[propane-1,3-diylbis(6-methoxy-1-benzothiene-5,2-diyl)]bis(4-oxobutanoic acid), fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
7MX4
 
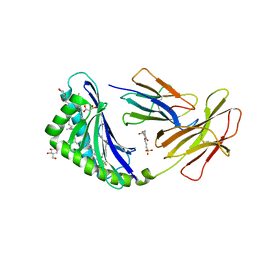 | | CD1c with antigen analogue 1 | | Descriptor: | (2S)-2,3-dihydroxypropyl hexadecanoate, (4S,8R,12S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl (1R,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl hydrogen (R)-phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cao, T.P, Shahine, A, Rossjohn, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
6LK2
 
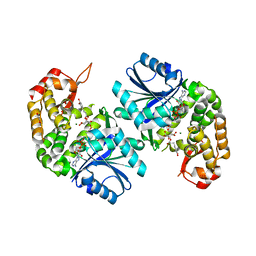 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+, NAD and chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 3-dehydroquinate synthase, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
7SD2
 
 | |
7MXF
 
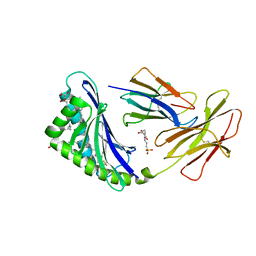 | | CD1c with antigen analogue 2 | | Descriptor: | (2S)-2,3-dihydroxypropyl hexadecanoate, 2,6-anhydro-1-deoxy-1-[(S)-hydroxy{[(4R,8S,12R,16R,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-D-glycero-D-galacto-heptitol, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cao, T.P, Shahine, A, Rossjohn, J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
4ZRB
 
 | |
6O4D
 
 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
7S99
 
 | | Room-temperature Human Hsp90a-NTD bound to N6M | | Descriptor: | Heat shock protein HSP 90-alpha, N-METHYL-9H-PURIN-6-AMINE | | Authors: | Stachowski, T.R, Vanarotti, M, Lopez, K, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8P8R
 
 | | Ex vivo Ym1 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 3 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
7MXN
 
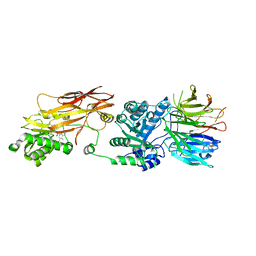 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
6LL9
 
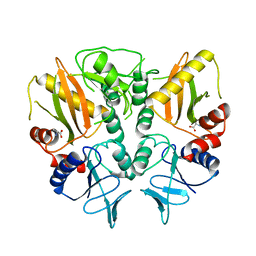 | |
6ULZ
 
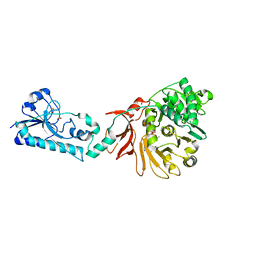 | | Adenylation domain of the initiation module of LgrA mutant P483M | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FORMIC ACID, ... | | Authors: | Chiche-Lapierre, C, Alonzo, D.A, Schmeing, T.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
7N0W
 
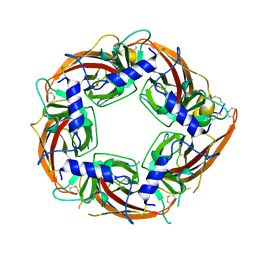 | |
7NP1
 
 | | Crystal Structure of the SARS-CoV-2 Receptor Binding Domain in Complex with Antibody ION-360 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin light chain, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Front Immunol, 12, 2021
|
|
7VL2
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with ethyl alpha-D-Glucoside | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-ethoxy-6-(hydroxymethyl)oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VDQ
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 7 | | Descriptor: | 2-[[7-[[2-fluoranyl-4-[3-(hydroxymethyl)pyrazol-1-yl]phenyl]amino]-1,6-naphthyridin-2-yl]-(1-methylpiperidin-4-yl)amino]ethanoic acid, Cyclin-dependent kinase 5 activator 1, p25, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7MX7
 
 | | PRMT5:MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
