5FMF
 
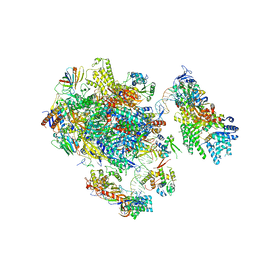 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8QSQ
 
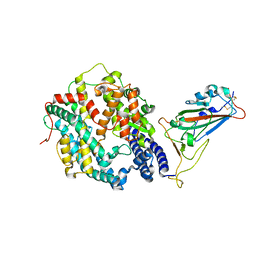 | |
7BLZ
 
 | | Red alga C.merolae Photosystem I | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Nelson, N, Klaiman, D, Hippler, M. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Red alga C.merolae Photosystem I
To Be Published
|
|
5FJ2
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((4-CHLORO-3-((METHYLAMINO)METHYL) PHENOXY)METHYL)QUINOLIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-10-06 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5GAD
 
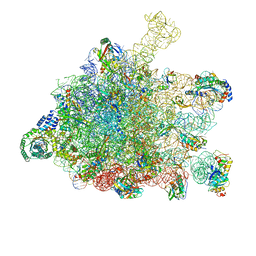 | | RNC-SRP-SR complex early state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
5G53
 
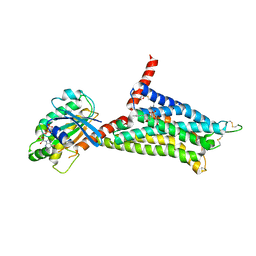 | | Structure of the adenosine A2A receptor bound to an engineered G protein | | Descriptor: | ADENOSINE RECEPTOR A2A, ENGINEERED DOMAIN OF HUMAN G ALPHA S LONG ISOFORM, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Carpenter, B, Nehme, R, Warne, T, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Adenosine A2A Receptor Bound to an Engineered G Protein
Nature, 536, 2016
|
|
8R8K
 
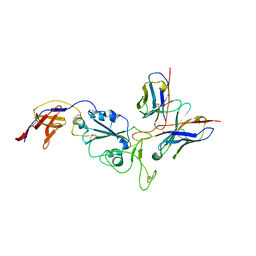 | | XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | Spike glycoprotein,Fibritin, XBB-4 Fab Heavy chain, XBB-4 Fab Light chain | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
5G6J
 
 | |
8R74
 
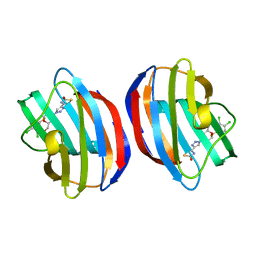 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-(2-oxidanyl-1,3-thiazol-4-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J.Med.Chem., 67, 2024
|
|
7ASN
 
 | |
5G6E
 
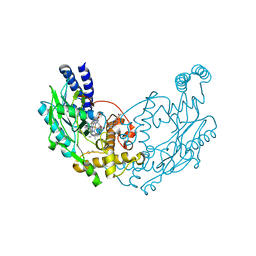 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 7-(((3-(Pyridin-3-yl)propyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-(((3-(PYRIDIN-3-YL)PROPYL)AMINO)METHYL)QUINOLIN-2-, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
7R15
 
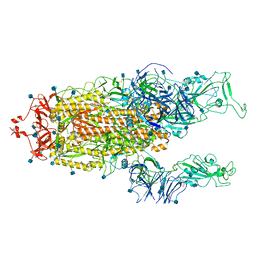 | | Alpha Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
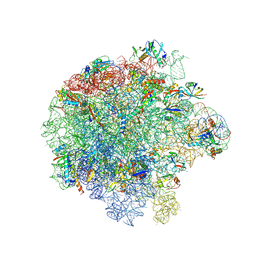
7ASM
 
 | |
5GAQ
 
 | | Cryo-EM structure of the Lysenin Pore | | Descriptor: | Lysenin | | Authors: | Savva, C.G, Bokori-Brown, M, Martin, T.G, Titball, R.W, Naylor, C.E, Basak, A.K. | | Deposit date: | 2016-01-05 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein
Nat Commun, 7, 2016
|
|
7B74
 
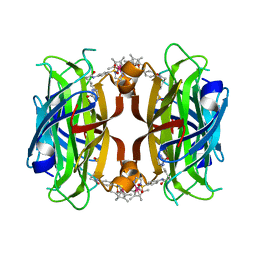 | | Chimeric Streptavidin With A Dimerization Domain For Artificial Transfer Hydrogenation | | Descriptor: | Streptavidin,Superoxide dismutase [Cu-Zn],Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spiers Memorial Lecture: Shielding the active site: a streptavidin superoxide-dismutase chimera as a host protein for asymmetric transfer hydrogenation.
Faraday Disc.Chem.Soc, 2023
|
|
7AU6
 
 | |
7ATN
 
 | |
7R10
 
 | | Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R5F
 
 | | Crystal structure of YTHDF2 with compound YLI_DF_012 | | Descriptor: | 5-azanyl-6-methyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7ATE
 
 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
7AU3
 
 | | Cytochrome c oxidase structure in F-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cytochrome c oxidase structure in F-state
To Be Published
|
|
5G6Q
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 7-(((5-((Methylamino)methyl)pyridin-3-yl)oxy)methyl) quinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[5-[(methylideneamino)methyl]pyridin-3-yl]oxymethyl]quinolin-2-amine, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
8QE8
 
 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
7R5L
 
 | | Crystal structure of YTHDF2 with compound YLI_DC1_015 | | Descriptor: | 3,6-dimethyl-2~{H}-1,2,4-triazin-5-one, SULFATE ION, YTH domain-containing family protein 2 | | Authors: | Nachawati, R, Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7R1A
 
 | | Furin Cleaved Alpha Variant SARS-CoV-2 Spike in complex with 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
