7WBS
 
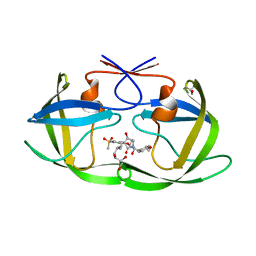 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
2OPP
 
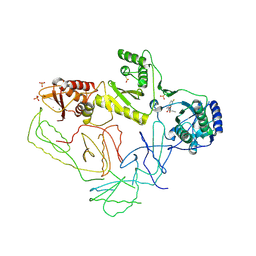 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
5OUK
 
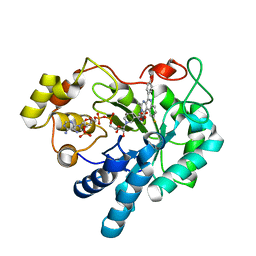 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
6PX0
 
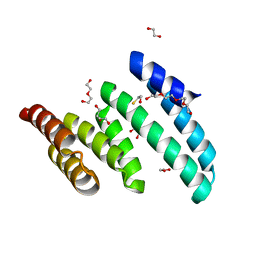 | |
4NZ6
 
 | | Steroid receptor RNA Activator (SRA) modification by the human Pseudouridine Synthase 1 (hPus1p): RNA binding, activity, and atomic model | | Descriptor: | 1,2-ETHANEDIOL, D-GLUTAMIC ACID, D-LYSINE, ... | | Authors: | Huet, T, Thore, S. | | Deposit date: | 2013-12-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid Receptor RNA Activator (SRA) Modification by the Human Pseudouridine Synthase 1 (hPus1p): RNA Binding, Activity, and Atomic Model
Plos One, 9, 2014
|
|
6DIF
 
 | | Wild-type HIV-1 protease in complex with tipranavir | | Descriptor: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DV4
 
 | | HIV-1 wild type protease with GRL-04315A, a tetrahydronaphthalene carboxamide with (R)-Boc-amine and (S)-hydroxyl functionalities as the P2 ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
6DJ7
 
 | | HIV-1 protease with mutation L76V in complex with GRL-5010 (gem-difluoro-bis-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
5OU0
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
2E39
 
 | | Crystal structure of the CN-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6DIL
 
 | | HIV-1 protease with single mutation L76V in complex with tipranavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DJ1
 
 | | Wild-type HIV-1 protease in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
2E3A
 
 | | Crystal structure of the NO-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRIC OXIDE, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6DH5
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DGZ
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH1
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH0
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH8
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
9MEA
 
 | |
6DGX
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH6
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
9JZB
 
 | | PfDXR - Mn2+ - MAMK251 ternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
9JZE
 
 | | PfDXR - Mn2+ - NADPH - MAMK431 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-10-14 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Diverse Binding Modes Explain the Nanomolar Levels of Inhibitory Activities Against 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase from Plasmodium falciparum Exhibited by Reverse Hydroxamate Analogs of Fosmidomycin with Varying N -Substituents.
Molecules, 30, 2024
|
|
