8GSX
 
 | |
6TXT
 
 | |
7OA8
 
 | |
7OA7
 
 | | PilC minor pilin of Streptococcus sanguinis 2908 type IV pili | | Descriptor: | CALCIUM ION, PilC minor pilin, SODIUM ION | | Authors: | Sheppard, D, Pelicic, V. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of a glycan-binding complex of minor pilins completes the analysis of Streptococcus sanguinis type 4 pili subunits.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DX4
 
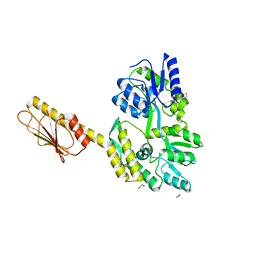 | |
5ZFR
 
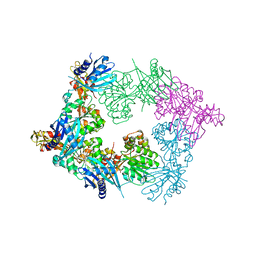 | | Crystal structure of PilB, an extension ATPase motor of Type IV pilus, from Geobacter sulfurreducens | | Descriptor: | PHOSPHATE ION, Type IV pilus biogenesis ATPase PilB, ZINC ION | | Authors: | Thakur, K.G, Solanki, V, Kapoor, S. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of Type IVa pilus extension and retraction ATPase motors
FEBS J., 285, 2018
|
|
5ZFQ
 
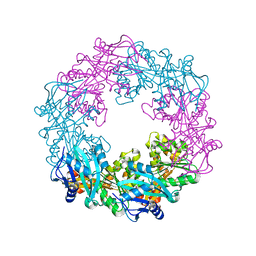 | |
7W65
 
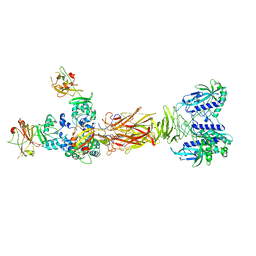 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with secreted protein TcpF | | Descriptor: | Toxin coregulated pilus biosynthesis protein F, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
5G2F
 
 | |
5G24
 
 | |
5G25
 
 | | Type IV-like pilin TTHA1218 from Thermus thermophilus | | Descriptor: | GLYCEROL, TYPE-IV LIKE COMPETENCE PILIN TTHA1218 | | Authors: | Karuppiah, V, Derrick, J.P. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-14 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Type Iv Pilins from Thermus Thermophilus Demonstrate Similarities with Type II Secretion System Pseudopilins
J.Struct.Biol., 196, 2016
|
|
5G23
 
 | |
7W64
 
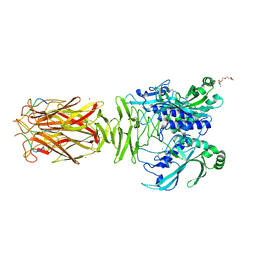 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with N-terminal peptide fragment of TcpF | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W63
 
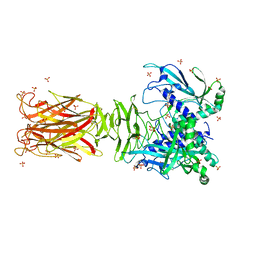 | | Crystal structure of minor pilin TcpB from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
6MIC
 
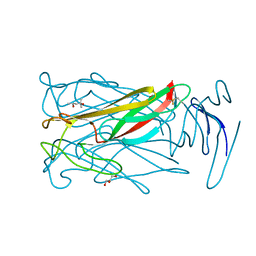 | |
1Q5F
 
 | | NMR Structure of Type IVb pilin (PilS) from Salmonella typhi | | Descriptor: | PilS | | Authors: | Xu, X.F, Tan, Y.W, Hackett, J, Zhang, M, Mok, Y.K. | | Deposit date: | 2003-08-07 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Type IVb Pilin from Salmonella typhi and Its Assembly into Pilus
J.Biol.Chem., 279, 2004
|
|
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
2M3K
 
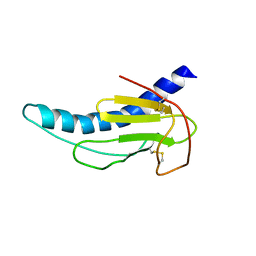 | |
6W6M
 
 | |
4IXJ
 
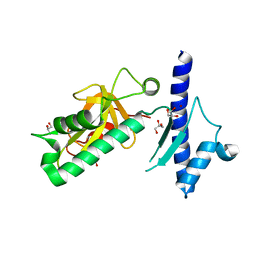 | | The structure of PilJ, a Type IV pilin from Clostridium difficile | | Descriptor: | Fimbrial protein (Pilin), GLYCEROL, ZINC ION | | Authors: | Piepenbrink, K.H, Sundberg, E.J, Snyder, G.A. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure of Clostridium difficile PilJ Exhibits Unprecedented Divergence from Known Type IV Pilins.
J.Biol.Chem., 289, 2014
|
|
3GN9
 
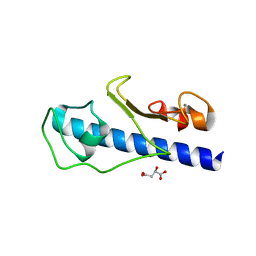 | | Crystal structure of the major pseudopilin from the type 2 secretion system of Vibrio vulnificus | | Descriptor: | CALCIUM ION, D-MALATE, Type II secretory pathway, ... | | Authors: | Korotkov, K.V, Gray, M.D, Kreger, A, Turley, S, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-03-16 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Calcium is essential for the major pseudopilin in the type 2 secretion system.
J.Biol.Chem., 284, 2009
|
|
2RET
 
 | | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the Type 2 Secretion System of Vibrio vulnificus | | Descriptor: | CHLORIDE ION, EpsJ, Pseudopilin EpsI, ... | | Authors: | Yanez, M.E, Korotkov, K.V, Abendroth, J, Hol, W.G.J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
J.Mol.Biol., 375, 2008
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7M0R
 
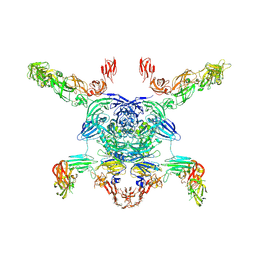 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
7M22
 
 | |
