9GD3
 
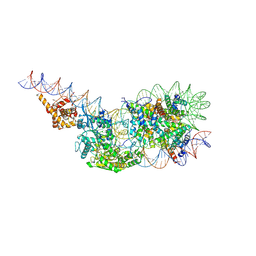 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
6RYR
 
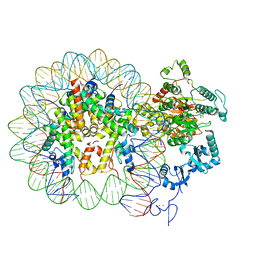 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6FTX
 
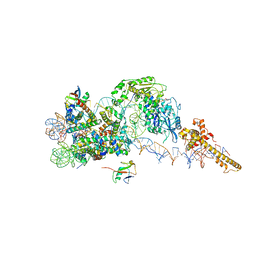 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
8UXQ
 
 | |
6G0L
 
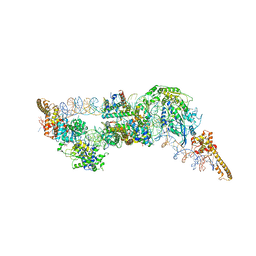 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
7TN2
 
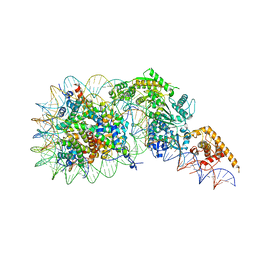 | | Composite model of a Chd1-nucleosome complex in the nucleotide-free state derived from 2.3A and 2.7A Cryo-EM maps | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6S47
 
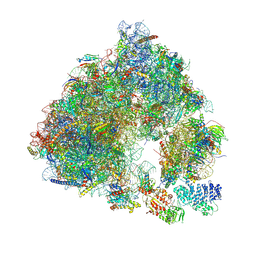 | | Saccharomyces cerevisiae 80S ribosome bound with ABCF protein New1 | | Descriptor: | 18S rRNA (1707-MER), 28S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Kasari, V, Pochopien, A.A, Margus, T, Murina, V, Turnbull, K, Zhou, Y, Nissan, T, Graf, M, Novacek, J, Atkinson, G.C, Johansson, M.J.O, Wilson, D.N, Hauryliuk, V. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A role for the Saccharomyces cerevisiae ABCF protein New1 in translation termination/recycling.
Nucleic Acids Res., 47, 2019
|
|
4FT4
 
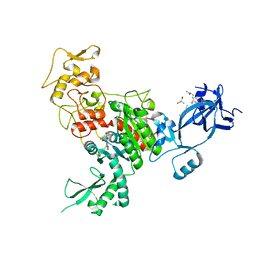 | |
7R76
 
 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
7R77
 
 | |
7R78
 
 | |
4FSX
 
 | |
6RYU
 
 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
7T02
 
 | |
4FT2
 
 | |
7UBU
 
 | |
3MWY
 
 | |
9NH8
 
 | | CHD1-nucleosome complex (anchored state) | | Descriptor: | ARGININE, Chromodomain-helicase-DNA-binding protein 1, DNA (158-MER), ... | | Authors: | James, A.M, Farnung, L. | | Deposit date: | 2025-02-24 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human CHD1 nucleosome recruitment and pausing.
Mol.Cell, 85, 2025
|
|
9MVY
 
 | | Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide | | Descriptor: | C49 ssDNA, DNA (cytosine-5)-methyltransferase 1, Histone H3.2, ... | | Authors: | Herle, G, Fang, J, Song, J. | | Deposit date: | 2025-01-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an Unfavorable de Novo DNA Methylation Complex of Plant Methyltransferase ZMET2.
J.Mol.Biol., 437, 2025
|
|
9N6K
 
 | | 2.88 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex with DNA-binding domain | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9N6H
 
 | | 2.54 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 1:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9N6I
 
 | | 2.61 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
5E4W
 
 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
