8IBK
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
6BS6
 
 | | SusG with mixed linkage amylosaccharide | | Descriptor: | ACETATE ION, Alpha-amylase SusG, CALCIUM ION, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for the flexible recognition of alpha-glucan substrates by Bacteroides thetaiotaomicron SusG.
Protein Sci., 27, 2018
|
|
8IDA
 
 | | Overall structure of the LAT1-4F2hc bound with tyrosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-02-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tyrosine
To Be Published
|
|
3OLG
 
 | | Structures of human pancreatic alpha-amylase in complex with acarviostatin III03 | | Descriptor: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes.
J.Struct.Biol., 174, 2011
|
|
3OLE
 
 | | Structures of human pancreatic alpha-amylase in complex with acarviostatin II03 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes.
J.Struct.Biol., 174, 2011
|
|
3OLI
 
 | | Structures of human pancreatic alpha-amylase in complex with acarviostatin IV03 | | Descriptor: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes.
J.Struct.Biol., 174, 2011
|
|
4X0N
 
 | |
4MAZ
 
 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
7LV6
 
 | | The structure of MalL mutant enzyme S536R from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hamill, C.J, Prentice, E.J, Bahl, C.D, Truebridge, I.S, Arcus, V.L. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Urea binding to guide rational design of mutations that influence enzyme dynamics
To Be Published
|
|
4MB1
 
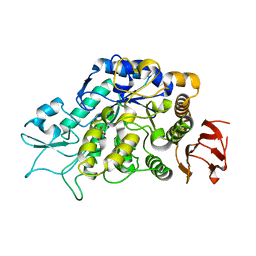 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4LQ1
 
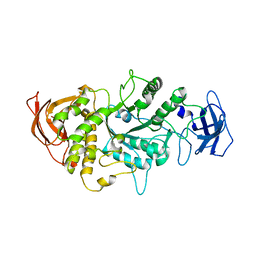 | |
4M8U
 
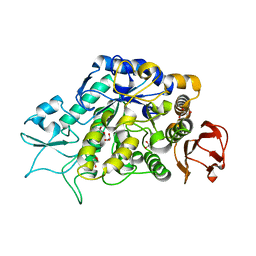 | | The Structure of MalL mutant enzyme V200A from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
8J8M
 
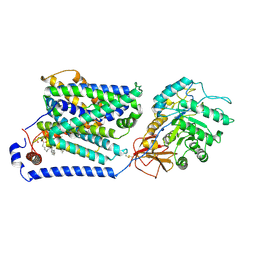 | | Overall structure of the LAT1-4F2hc bound with tryptophan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tryptophan
To Be Published
|
|
8J8L
 
 | | Overall structure of the LAT1-4F2hc bound with L-dopa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-DIHYDROXYPHENYLALANINE, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with L-dopa
To Be Published
|
|
4X9Y
 
 | |
7ML5
 
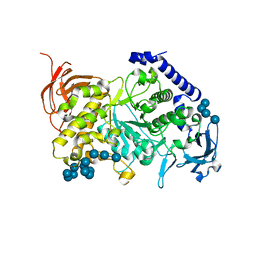 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
4M56
 
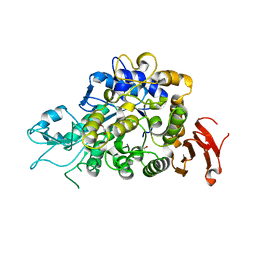 | | The Structure of Wild-type MalL from Bacillus subtilis | | Descriptor: | D-glucose, GLYCEROL, Oligo-1,6-glucosidase 1, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
7U3D
 
 | | Structure of S. venezuelae GlgX-c-di-GMP-acarbose complex (4.6) | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U3B
 
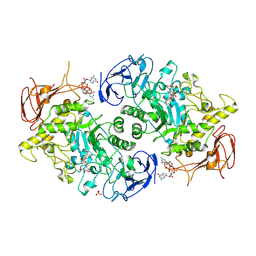 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | Authors: | Schumacher, M.A, Tschowri, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U39
 
 | |
8OR6
 
 | |
8ORP
 
 | | Crystal structure of Drosophila melanogaster alpha-amylase in complex with the inhibitor acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of Drosophila melanogaster alpha-Amylase.
Molecules, 28, 2023
|
|
7NF8
 
 | | Ovine (b0,+AT-rBAT)2 hetero-tetramer, asymmetric unit, rigid-body fitted | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF6
 
 | | Ovine b0,+AT-rBAT heterodimer | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7U3A
 
 | | Structure of the Streptomyces venezuelae GlgX-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
