2ISL
 
 | | BluB bound to reduced flavin (FMNH2) and molecular oxygen. (clear crystal form) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB, OXYGEN MOLECULE | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2MIH
 
 | |
2ISK
 
 | | BluB bound to flavin anion (charge transfer complex) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2MBT
 
 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
2J4D
 
 | | Cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, CRYPTOCHROME DASH, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Klar, T, Pokorny, R, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryptochrome 3 from Arabidopsis Thaliana: Structural and Functional Analysis of its Complex with a Folate Light Antenna
J.Mol.Biol., 366, 2007
|
|
2M9K
 
 | | RBPMS2-Nter | | Descriptor: | RNA-binding protein with multiple splicing 2 | | Authors: | Yang, Y. | | Deposit date: | 2013-06-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity.
Nucleic Acids Res., 42, 2014
|
|
2MIF
 
 | |
1SUQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1L6T
 
 | |
1TLF
 
 | |
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
2NTG
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OGG
 
 | | Structure of B. subtilis trehalose repressor (TreR) effector binding domain | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Rezacova, P, Krejcirikova, V, Borek, D, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly.
Proteins, 69, 2007
|
|
2NZ0
 
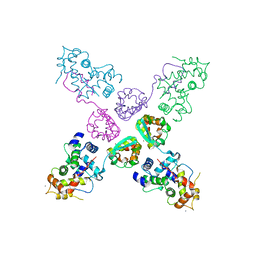 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
1ZAK
 
 | |
2JA1
 
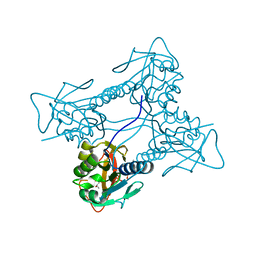 | | Thymidine kinase from B. cereus with TTP bound as phosphate donor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2OAU
 
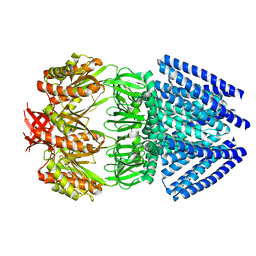 | | Mechanosensitive Channel of Small Conductance (MscS) | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Rees, D.C, Bass, R.B, Steinbacher, S, Strop, P, Barclay, M.T. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
CURRENT TOPICS IN MEMBRANES, 58, 2007
|
|
1S6Q
 
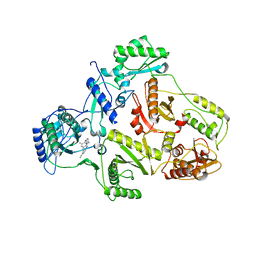 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1M5S
 
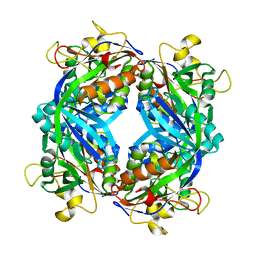 | | Formylmethanofuran:tetrahydromethanopterin fromyltransferase from Methanosarcina barkeri | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
2IF7
 
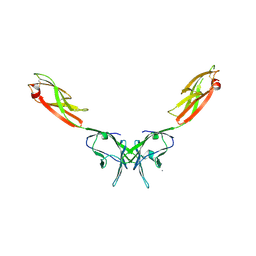 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
2J7A
 
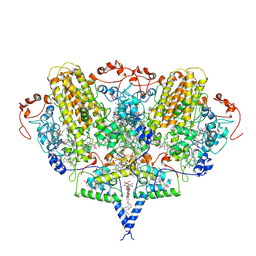 | | Crystal structure of cytochrome c nitrite reductase NrfHA complex from Desulfovibrio vulgaris | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE NRFA, ... | | Authors: | Rodrigues, M.L, Oliveira, T.F, Pereira, I.A.C, Archer, M. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of the Membrane-Bound Cytochrome C Quinol Dehydrogenase Nrfh Reveals Novel Haem Coordination.
Embo J., 25, 2006
|
|
2BOK
 
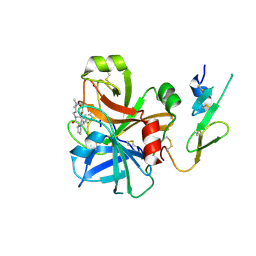 | | Factor Xa - cation | | Descriptor: | COAGULATION FACTOR X, SODIUM ION, [AMINO (4-{(3AS,4R,8AS,8BR)-1,3-DIOXO-2- [3-(TRIMETHYLAMMONIO) PROPYL]DECAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL}PHENYL) METHYLENE]AMMONIUM | | Authors: | Morgenthaler, M, Schaerer, K, Paulini, R, Obst-Sander, U, Banner, D.W, Schlatter, D, Benz, J, Stihle, M, Diederich, F. | | Deposit date: | 2005-04-12 | | Release date: | 2005-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Quantification of Cation-Pi Interactions in Protein-Ligand Complexes: Crystal-Structure Analysis of Factor Xa Bound to a Quaternary Ammonium Ion Ligand
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1LPV
 
 | | DROSOPHILA MELANOGASTER DOUBLESEX (DSX), NMR, 18 STRUCTURES | | Descriptor: | Doublesex protein, ZINC ION | | Authors: | Zhu, L, Wilken, J, Phillips, N, Narendra, U, Chan, G, Stratton, S, Kent, S, Weiss, M.A. | | Deposit date: | 2002-05-08 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers.
Genes Dev., 14, 2000
|
|
1BTU
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4R)-1-TOLUENESULPHONYL-3-ETHYL-AZETIDIN-2-ONE-4-CARBOXYLIC ACID | | Descriptor: | (3R)-3-ethyl-N-[(4-methylphenyl)sulfonyl]-L-aspartic acid, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of elastase by N-sulfonylaryl beta-lactams: anatomy of a stable acyl-enzyme complex.
Biochemistry, 37, 1998
|
|
