4YMY
 
 | | Crystal structure of mutant nitrobindin M75A/H76L/Q96C/M148L/H158A (NB11) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Himiyama, T, Tachikawa, K, Oohora, K, Onoda, A, Hayashi, T. | | Deposit date: | 2015-03-08 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Highly Active Biohybrid Catalyst for Olefin Metathesis in Water: Impact of a Hydrophobic Cavity in a beta-Barrel Protein
Acs Catalysis, 5, 2015
|
|
6YZU
 
 | | Carborane nido-pentyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane nido-pentyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
4GNR
 
 | | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine | | Descriptor: | ABC transporter substrate-binding protein-branched chain amino acid transport, CHLORIDE ION, ISOLEUCINE, ... | | Authors: | Halavaty, A.S, Kudritska, M, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine
To be Published
|
|
1K4P
 
 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with zinc ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, SULFATE ION, ZINC ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
1OAI
 
 | |
1Y55
 
 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | Descriptor: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5D14
 
 | |
2WYT
 
 | | 1.0 A resolution structure of L38V SOD1 mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
6Q49
 
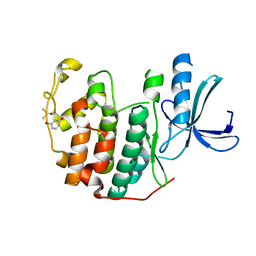 | | CDK2 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7FBM
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing dibutylhydroxytoluene (BHT) | | Descriptor: | 2,6-ditert-butyl-4-methyl-phenol, ACETIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing dibutylhydroxytoluene (BHT)
To Be Published
|
|
6C3T
 
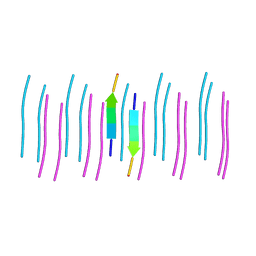 | |
1JFB
 
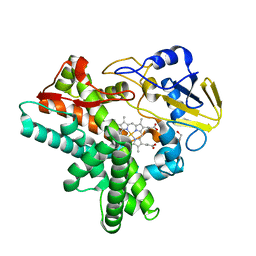 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8DA5
 
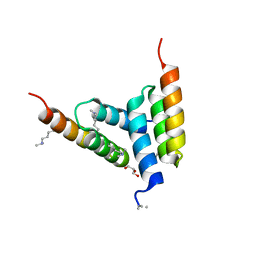 | | Coevolved affibody-Z domain pair LL1.c4 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein A, affibody LL1.FIVM | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
7FD7
 
 | | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid | | Descriptor: | Fatty acid-binding protein, heart, PENTAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid
To Be Published
|
|
6RD2
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3FYM
 
 | | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Xu, L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2009-01-22 | | Release date: | 2010-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus
To be Published
|
|
6I3B
 
 | |
3AUB
 
 | | A simplified BPTI variant stabilized by the A14G and A38V substitutions | | Descriptor: | Bovine Pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
4LN2
 
 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
2GGC
 
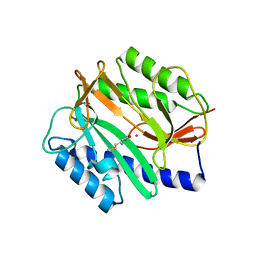 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHIONINE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
6ZFJ
 
 | |
7BR5
 
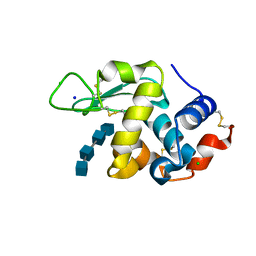 | | Lysozyme-sugar complex in H2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2P5K
 
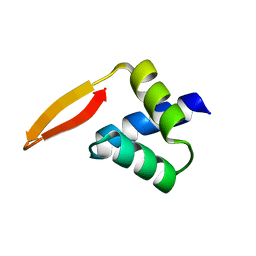 | | Crystal structure of the N-terminal domain of AhrC | | Descriptor: | Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1R2M
 
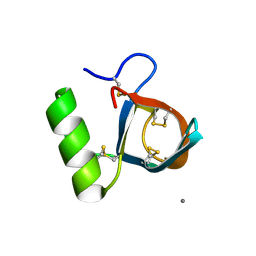 | | Atomic resolution structure of the HFBII hydrophobin: a self-assembling amphiphile | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Paananen, A, Askolin, S, Nakari-Setala, T, Parkkinen, T, Penttila, M, Linder, M.B, Rouvinen, J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile.
J.Biol.Chem., 279, 2004
|
|
6FVI
 
 | |
