2SGQ
 
 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
5IQ8
 
 | |
2G9F
 
 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GFP
 
 | | Structure of the Multidrug Transporter EmrD from Escherichia coli | | Descriptor: | Multidrug resistance protein D | | Authors: | Yin, Y, He, X, Szewczyk, P, Nguyen, T, Chang, G. | | Deposit date: | 2006-03-22 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the multidrug transporter EmrD from Escherichia coli
Science, 312, 2006
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
2STA
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-10 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2RMX
 
 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | Descriptor: | NKG2-A/NKG2-B type II integral membrane protein, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of the two NKG2A immunoreceptor tyrosine-based inhibitory motifs (ITIMs) by the C-terminal SH2 domain of protein tyrosine phosphatase SHP-1
To be Published
|
|
2RNO
 
 | |
2ROB
 
 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2GHH
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | NITRIC OXIDE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2TMN
 
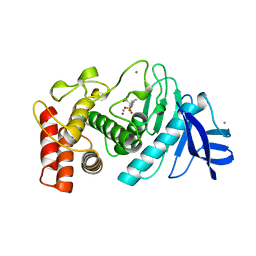 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
2RPB
 
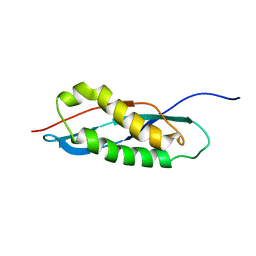 | |
5I38
 
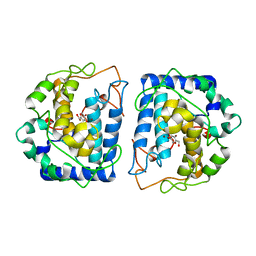 | | Crystal Structure of tyrosinase from Bacillus megaterium with inhibitor kojic acid in the active site | | Descriptor: | 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, COPPER (II) ION, Tyrosinase | | Authors: | Kanteev, M, Goldfeder, M, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2016-02-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The unravelling of the complex pattern of tyrosinase inhibition.
Sci Rep, 6, 2016
|
|
2RQX
 
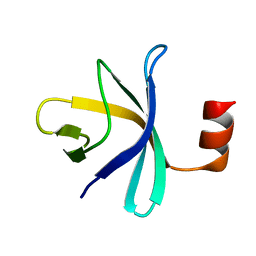 | |
2TPS
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PROTEIN (THIAMIN PHOSPHATE SYNTHASE), PYROPHOSPHATE 2-, ... | | Authors: | Chiu, H.-J, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
Biochemistry, 38, 1999
|
|
2RR3
 
 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | Descriptor: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | Authors: | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
5I4B
 
 | |
2GCL
 
 | | Structure of the Pob3 Middle domain | | Descriptor: | CHLORIDE ION, Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE), ... | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
2RVB
 
 | |
5I52
 
 | |
2SN3
 
 | | STRUCTURE OF SCORPION TOXIN VARIANT-3 AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SCORPION NEUROTOXIN (VARIANT 3) | | Authors: | Zhao, B, Carson, M, Ealick, S.E, Bugg, C.E. | | Deposit date: | 1992-02-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of scorpion toxin variant-3 at 1.2 A resolution.
J.Mol.Biol., 227, 1992
|
|
2TIR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT ESCHERICHIA COLI THIOREDOXIN IN WHICH LYSINE 36 IS REPLACED BY GLUTAMIC ACID | | Descriptor: | COPPER (II) ION, THIOREDOXIN | | Authors: | Nikkola, M, Gleason, F.K, Fuchs, J.A, Eklund, H. | | Deposit date: | 1993-01-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of a mutant Escherichia coli thioredoxin in which lysine 36 is replaced by glutamic acid.
Biochemistry, 32, 1993
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2GEY
 
 | | Crystal Structure of AclR a putative hydroxylase from Streptomyces galilaeus | | Descriptor: | AclR protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Beinker, P, Lohkamp, B, Schneider, G. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of SnoaL2 and AclR: two putative hydroxylases in the biosynthesis of aromatic polyketide antibiotics
J.Mol.Biol., 359, 2006
|
|
