3ZD8
 
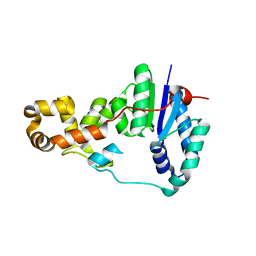 | | Potassium bound structure of E. coli ExoIX in P1 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
1T03
 
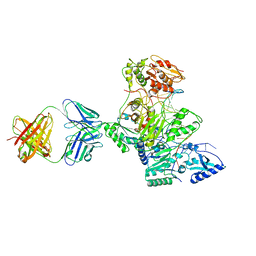 | | HIV-1 reverse transcriptase crosslinked to tenofovir terminated template-primer (complex P) | | Descriptor: | MAGNESIUM ION, POL polyprotein, Synthetic oligonucleotide primer, ... | | Authors: | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | Deposit date: | 2004-04-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
2KBZ
 
 | |
1GID
 
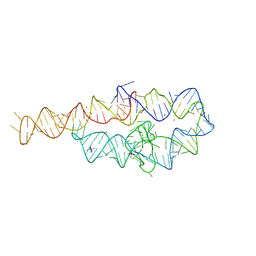 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
3GOM
 
 | | Barium bound to the Holliday junction sequence d(TCGGCGCCGA)4 | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION | | Authors: | Naseer, A, Cardin, C.J. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2CS1
 
 | |
8ASO
 
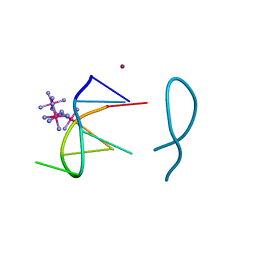 | | Nickel(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), NICKEL (II) ION | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8ASM
 
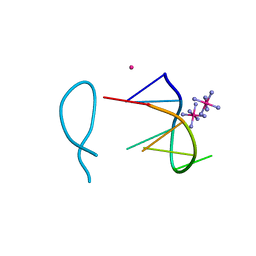 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
2ZIE
 
 | | Crystal Structure of TTHA0409, Putatative DNA Modification Methylase from Thermus thermophilus HB8- Selenomethionine derivative | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
2IUW
 
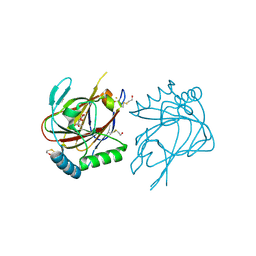 | | Crystal structure of human ABH3 in complex with iron ion and 2- oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ALKYLATED REPAIR PROTEIN ALKB HOMOLOG 3, BETA-MERCAPTOETHANOL, ... | | Authors: | Sundheim, O, Vagbo, C.B, Bjoras, M, deSousa, M.M.L, Talstad, V, Aas, P.A, Drablos, F, Krokan, H.E, Tainer, J.A, Slupphaug, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Abh3 Structure and Key Residues for Oxidative Demethylation to Reverse DNA/RNA Damage.
Embo J., 25, 2006
|
|
7ELN
 
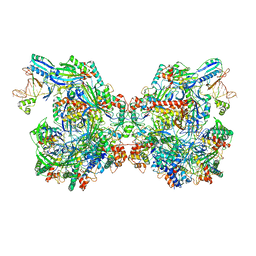 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
8BAG
 
 | |
8BAE
 
 | |
8BAF
 
 | |
454D
 
 | | INTERCALATION AND MAJOR GROOVE RECOGNITION IN THE 1.2 A RESOLUTION CRYSTAL STRUCTURE OF RH[ME2TRIEN]PHI BOUND TO 5'-G(5IU)TGCAAC-3' | | Descriptor: | 5'-D(*GP*(5IU)P*TP*GP*CP*AP*AP*C)-3', DELTA-ALPHA-RH[2R,9R-DIAMINO-4,7-DIAZADECANE]9,10-PHENANTHRENEQUINONE DIIMINE | | Authors: | Kielkopf, C.L, Erkkila, K.E, Hudson, B.P, Barton, J.K, Rees, D.C. | | Deposit date: | 1999-03-03 | | Release date: | 2000-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a photoactive rhodium complex intercalated into DNA.
Nat.Struct.Biol., 7, 2000
|
|
2N7D
 
 | |
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
8CLJ
 
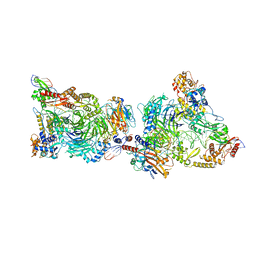 | | TFIIIC TauB-DNA dimer | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
1RZ9
 
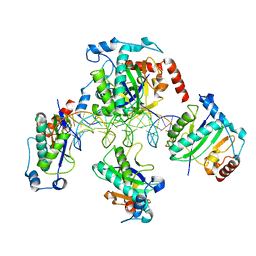 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
2JPC
 
 | |
2GCL
 
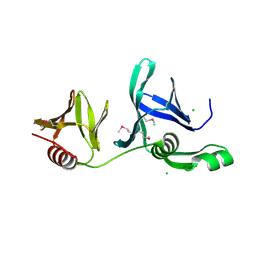 | | Structure of the Pob3 Middle domain | | Descriptor: | CHLORIDE ION, Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2ZIG
 
 | | Crystal Structure of TTHA0409, Putative DNA Modification Methylase from Thermus thermophilus HB8 | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
1J5O
 
 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2WIZ
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WJ0
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, COBALT HEXAMMINE(III), HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
