1DBM
 
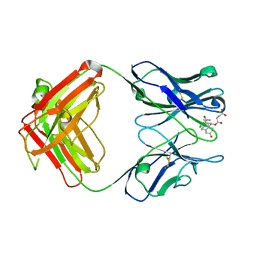 | |
1DBJ
 
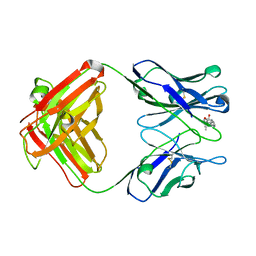 | |
1EAP
 
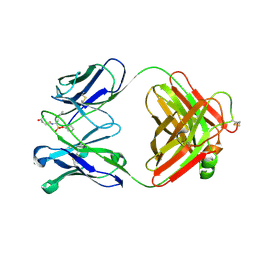 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
5JMO
 
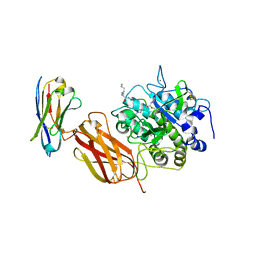 | |
8QXC
 
 | | Crystal structure of antibody Fab MIL-3 with PenG-Lys | | Descriptor: | (2R,4S)-2-[(1R)-2-[[(5S)-5-acetamido-6-oxidanyl-6-oxidanylidene-hexyl]amino]-2-oxidanylidene-1-(2-phenylethanoylamino)ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Heavy chain of FAB MIL-3, Light chain of FAB MIL-3 | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent penicillin-protein conjugates elicit anti-drug antibodies that are clonally and functionally restricted.
Nat Commun, 15, 2024
|
|
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
3AUV
 
 | | Predicting Amino Acid Preferences in the Complementarity Determining Regions of an Antibody-Antigen Recognition Interface | | Descriptor: | sc-dsFv derived from the G6-Fab | | Authors: | Yu, C.M, Peng, H.P, Chen, I.C, Lee, Y.C, Chen, J.B, Tsai, K.C, Chen, C.T, Chang, J.Y, Yang, E.W, Hsu, P.C, Jian, J.W, Hsu, H.J, Chang, H.J, Hsu, W.L, Huang, K.F, Ma, A.C, Yang, A.S. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface
Plos One, 7, 2012
|
|
2G75
 
 | | Crystal Structure of anti-SARS m396 Antibody | | Descriptor: | IGG Heavy Chain, IGG Light Chain | | Authors: | Prabakaran, P, Gan, J.H, Feng, Y, Zhu, Z.Y, Xiao, X.D, Ji, X, Dimitrov, D.S. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody.
J.Biol.Chem., 281, 2006
|
|
4RFO
 
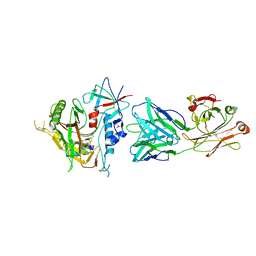 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
6KYZ
 
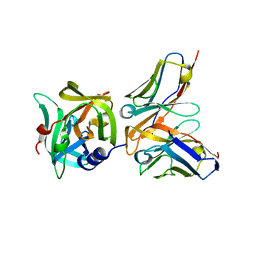 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
6LDX
 
 | |
6LDY
 
 | |
6LDW
 
 | |
1CS9
 
 | | SOLUTION STRUCTURE OF CGGIRGERA IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
6GV4
 
 | | High-resolution Cryo-EM of Fab-labeled human parechovirus 3 | | Descriptor: | AT12-015 antibody variable heavy, AT12-015 antibody variable light, RNA (5'-R(*UP*GP*GP*UP*AP*UP*UP*U)-3'), ... | | Authors: | Domanska, A, Flatt, J.W, Jukonen, J.J.J, Geraets, J.A, Butcher, S.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.
J.Virol., 93, 2019
|
|
6QFC
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | DIMETHYL SULFOXIDE, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5BV7
 
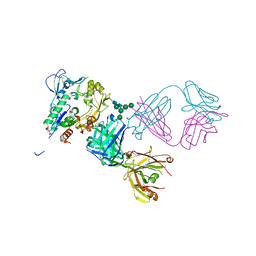 | | Crystal structure of human LCAT (L4F, N5D) in complex with Fab of an agonistic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 27C3 heavy chain, 27C3 light chain, ... | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Agonistic Human Antibodies Binding to Lecithin-Cholesterol Acyltransferase Modulate High Density Lipoprotein Metabolism.
J.Biol.Chem., 291, 2016
|
|
43CA
 
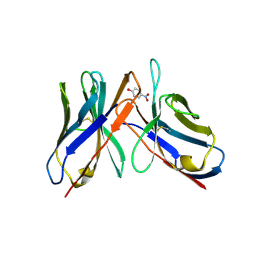 | |
43C9
 
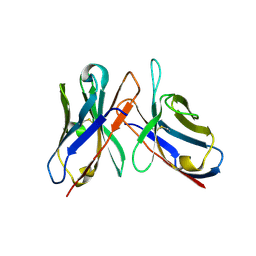 | |
1XF2
 
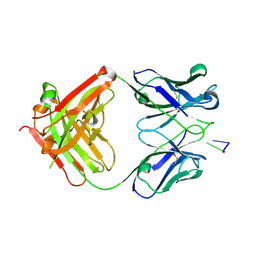 | | Structure of Fab DNA-1 complexed with dT3 | | Descriptor: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
2H1P
 
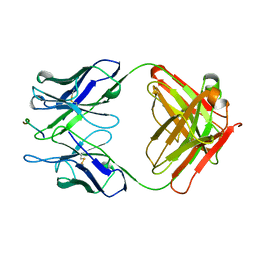 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1CW8
 
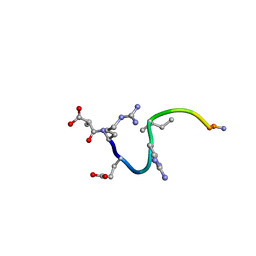 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO (mA-R)REGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 6 STRUCTURES | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, T. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
2XEG
 
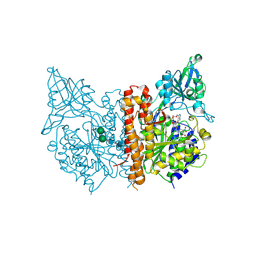 | | Human glutamate carboxypeptidase II in complex with Antibody- Recruiting Molecule ARM-P4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, A.X, Murelli, R.P, Barinka, C, Michel, J, Cocleaza, A, Jorgensen, W.L, Lubkowski, J, Spiegel, D.A. | | Deposit date: | 2010-05-14 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A Remote Arene-Binding Site on Prostate Specific Membrane Antigen Revealed by Antibody-Recruiting Small Molecules.
J.Am.Chem.Soc., 132, 2010
|
|
