3FSU
 
 | | Crystal Structure of Escherichia coli Methylenetetrahydrofolate Reductase Double Mutant Phe223LeuGlu28Gln complexed with methyltetrahydrofolate | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli.
Biochemistry, 48, 2009
|
|
4JFW
 
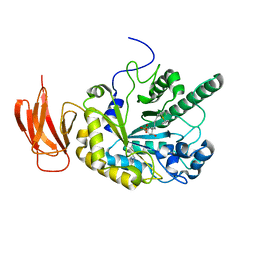 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor (2S,3S,4R,5S)-2-[N-(propylferrocene)]aminoethyl-5-methylpyrrolidine-3,4-diol | | Descriptor: | (3alpha)-[3-({2-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]ethyl}amino)propyl]ferrocene, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4O2W
 
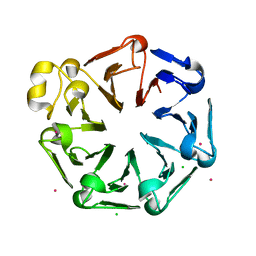 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
4JGN
 
 | |
4O3X
 
 | |
4JH7
 
 | | Crystal Structure of FosB from Bacillus cereus with Manganese and L-Cysteine-Fosfomycin Product | | Descriptor: | (2R)-2-azanyl-3-[(1R,2S)-2-oxidanyl-1-phosphono-propyl]sulfanyl-propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
4JI3
 
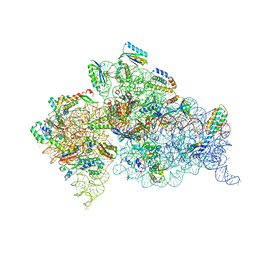 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy, F.V, Murphy, E.L, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JKG
 
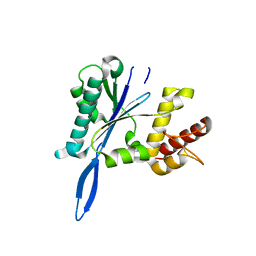 | | Open and closed forms of mixed T1789P+R1865A and R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3PHO
 
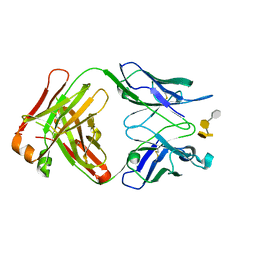 | | Crystal structure of S64-4 in complex with PSBP | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose, S64-4 Fab (IgG1) heavy chain, S64-4 Fab (IgG1) light chain | | Authors: | Evans, D.W, Evans, S.V. | | Deposit date: | 2010-11-04 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia.
Glycobiology, 21, 2011
|
|
3G3X
 
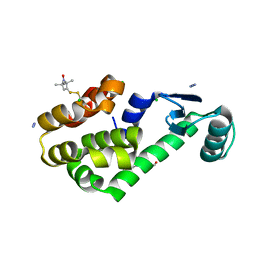 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 100 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
4NRZ
 
 | |
3FW2
 
 | | C-terminal domain of putative thiol-disulfide oxidoreductase from Bacteroides thetaiotaomicron. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, thiol-disulfide oxidoreductase | | Authors: | Osipiuk, J, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-16 | | Release date: | 2009-01-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystal structure of C-terminal domain of putative thiol-disulfide oxidoreductase from Bacteroides thetaiotaomicron.
To be Published
|
|
2OMM
 
 | |
3FWJ
 
 | | Ferric camphor bound Cytochrome P450cam containing a selenocysteine as the 5th heme ligand, orthorombic crystal form | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Schlichting, I, von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OMV
 
 | | Crystal structure of InlA S192N Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OQE
 
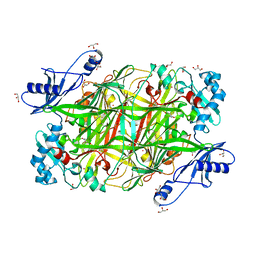 | |
3G77
 
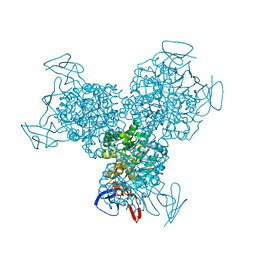 | | Bacterial cytosine deaminase V152A/F316C/D317G mutant | | Descriptor: | Cytosine deaminase, FE (III) ION | | Authors: | Stoddard, B, Zhao, L. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo.
Cancer Res., 69, 2009
|
|
4O0I
 
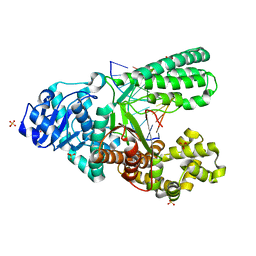 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA | | Descriptor: | 5'-D(*C*(1TW)P*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', DNA polymerase I, GLYCEROL, ... | | Authors: | Jiang, S, Gan, J, Sun, H, Huang, Z. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA
TO BE PUBLISHED
|
|
4WL9
 
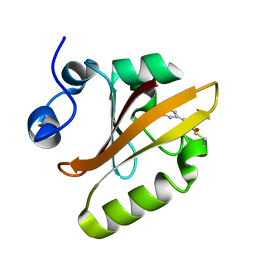 | |
4O1H
 
 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
3G96
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3PWZ
 
 | | Crystal structure of an Ael1 enzyme from Pseudomonas putida | | Descriptor: | Shikimate dehydrogenase 3 | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural and mechanistic analysis of a novel class of shikimate dehydrogenases: evidence for a conserved catalytic mechanism in the shikimate dehydrogenase family.
Biochemistry, 50, 2011
|
|
4K1O
 
 | |
2AWH
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-01 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
3PRD
 
 | |
