6VMF
 
 | | Crystal structure of the Y766F mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SULFATE ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
8ECP
 
 | |
4ZV1
 
 | |
8HPE
 
 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
8HKN
 
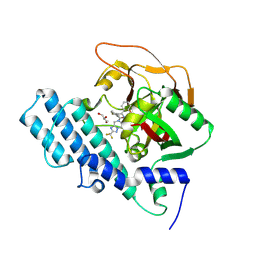 | |
4ZV2
 
 | |
6VO5
 
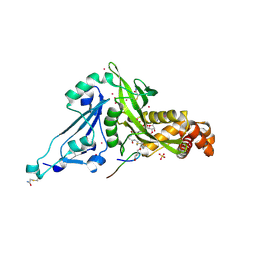 | | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4 | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4, ... | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
to be published
|
|
8HLQ
 
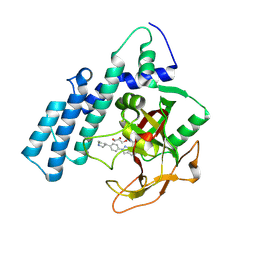 | |
4ZX4
 
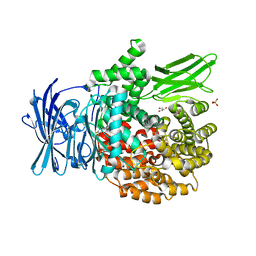 | |
8F5Q
 
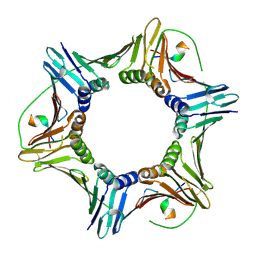 | | Crystal structure of human PCNA in complex with the PIP box of FBH1 | | Descriptor: | F-box DNA helicase 1, Proliferating cell nuclear antigen | | Authors: | Liu, J, Chaves-Arquero, B, Wei, P, Tencer, H, Zhang, G, Blanco, F, Kutateladze, T. | | Deposit date: | 2022-11-15 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insight into the PCNA-binding mode of FBH1.
Structure, 31, 2023
|
|
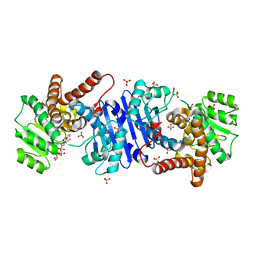
8HR6
 
 | |
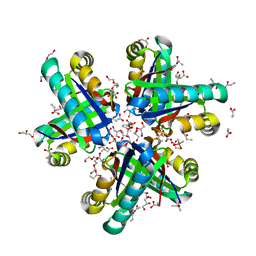
8EIF
 
 | |
8HLJ
 
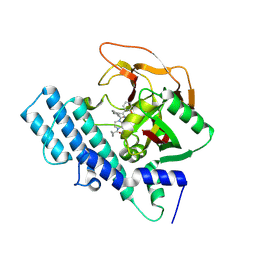 | |
6VP1
 
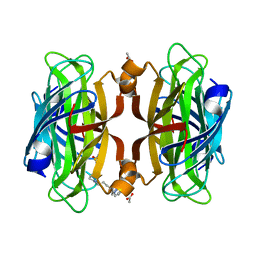 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
8HKO
 
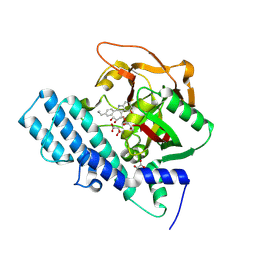 | |
8EM5
 
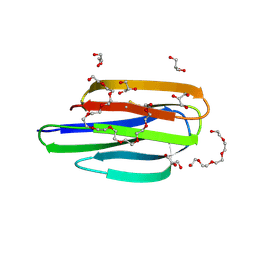 | | Mycobacterium thermoresistible MmpS5 | | Descriptor: | DODECAETHYLENE GLYCOL, GLYCEROL, IODIDE ION, ... | | Authors: | Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Mycobacterium thermoresistibile MmpS5 reveals a conserved disulfide bond across mycobacteria.
Metallomics, 16, 2024
|
|
4ZSJ
 
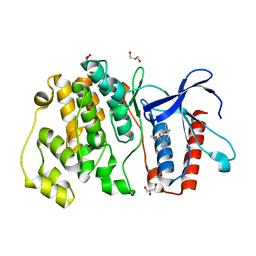 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chloro-3-methylphenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8HKS
 
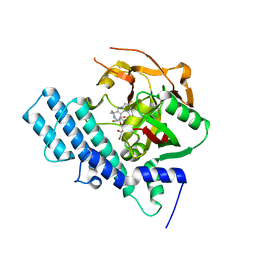 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
5KOW
 
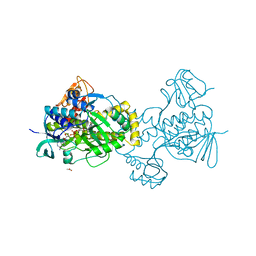 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
8HLR
 
 | |
4ZT2
 
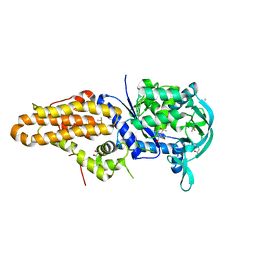 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)propane-1,3-diamine (Chem 1575) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, METHIONINE, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-05-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
8ECX
 
 | |
5KUT
 
 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
8EM3
 
 | |
4ZUK
 
 | | Structure ALDH7A1 complexed with NAD+ | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
