3NFF
 
 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | Descriptor: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
5MG8
 
 | | Crystal structure of the S.pombe Smc5/6 hinge domain | | Descriptor: | GLYCEROL, SULFATE ION, Structural maintenance of chromosomes protein 5, ... | | Authors: | Alt, A, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Specialized interfaces of Smc5/6 control hinge stability and DNA association.
Nat Commun, 8, 2017
|
|
5CD4
 
 | | The Type IE CRISPR Cascade complex from E. coli, with two assemblies in the asymmetric unit arranged back-to-back | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2015-07-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
1G8U
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATF5UCGCG):5-FORMYLURIDINE/ ADENOSINE BASE-PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | Authors: | Tsunoda, M, Karino, N, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2'-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers?
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1R05
 
 | | Solution Structure of Max B-HLH-LZ | | Descriptor: | Max protein | | Authors: | Sauv, S, Tremblay, L, Lavigne, P. | | Deposit date: | 2003-09-19 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
1G75
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATF5UCGCG): 5-FORMYLURIDINE/ ADENOSINE BASE-PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*(UFR)P*CP*GP*CP*G)-3', MAGNESIUM ION, POTASSIUM ION | | Authors: | Tsunoda, M, Karino, N, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2000-11-09 | | Release date: | 2001-02-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2'-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers?
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3F1W
 
 | |
6RYK
 
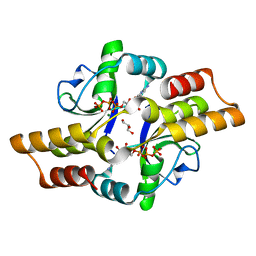 | | Crystal structure of the ParB-like protein PadC | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ParB-type DNA Segregation Proteins Are CTP-Dependent Molecular Switches.
Cell, 179, 2019
|
|
2XCT
 
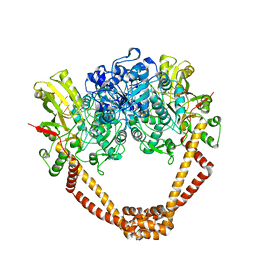 | | The twinned 3.35A structure of S. aureus Gyrase complex with Ciprofloxacin and DNA | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 5'-D(AP*GP*CP*CP*GP*TP*AP*G)-3', 5'-D(GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3', ... | | Authors: | Bax, B.D, Chan, P, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
1AKO
 
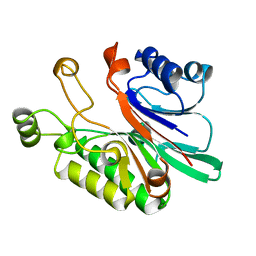 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
8K0S
 
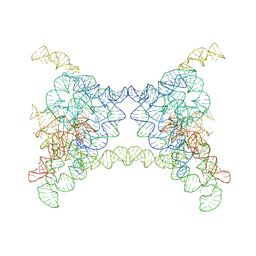 | |
3HA8
 
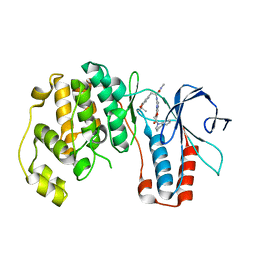 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/Compound 14b | | Descriptor: | Mitogen-activated protein kinase 14, N~2~-{4-[6-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)-1H-benzimidazol-1-yl]-6-ethoxy-1,3,5-triazin-2-yl}-3-(2,2-dimethyl-4H-1,3-benzodioxin-6-yl)-N-methyl-L-alaninamide | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
8K15
 
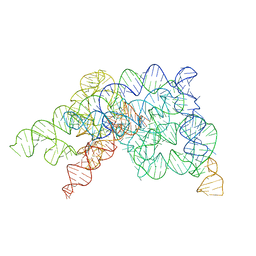 | |
3P87
 
 | | Structure of human PCNA bound to RNASEH2B PIP box peptide | | Descriptor: | Proliferating cell nuclear antigen, Ribonuclease H2 subunit B | | Authors: | Bubeck, D, Reijns, M.A, Graham, S.C, Astell, K.R, Jones, E.Y, Jackson, A.P. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | PCNA directs type 2 RNase H activity on DNA replication and repair substrates.
Nucleic Acids Res., 39, 2011
|
|
8K0P
 
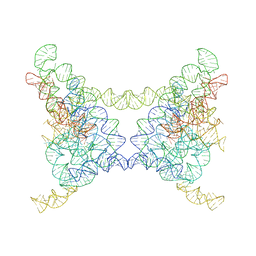 | |
8GYD
 
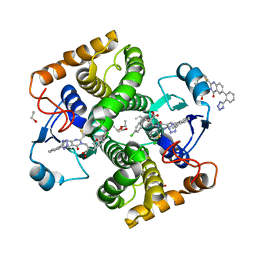 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
8W35
 
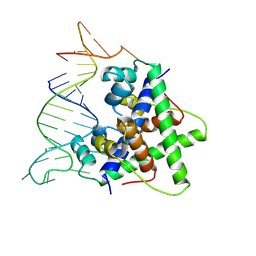 | | Aca2 from Pectobacterium phage ZF40 bound to RNA | | Descriptor: | Anti-CRISPR associated (Aca) protein, Aca2, IR2 and IR-RBS RNA | | Authors: | Wilkinson, M.E, Birkholz, N, Kimanius, D, Fineran, P.C. | | Deposit date: | 2024-02-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Phage anti-CRISPR control by an RNA- and DNA-binding helix-turn-helix protein.
Nature, 631, 2024
|
|
5OBZ
 
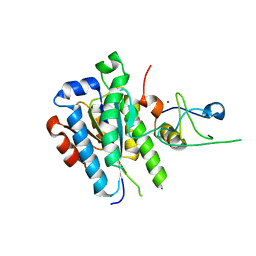 | | low resolution structure of the p34ct/p44ct minimal complex | | Descriptor: | Putative transcription factor, ZINC ION | | Authors: | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
2FBY
 
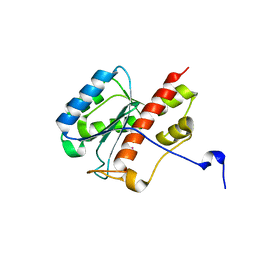 | | WRN exonuclease, Eu complex | | Descriptor: | EUROPIUM (III) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
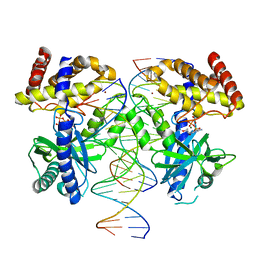
8SJ1
 
 | |
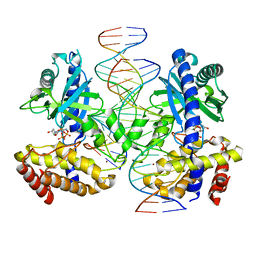
8SJ0
 
 | |
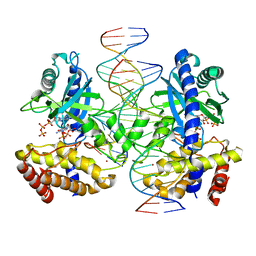
8SJ2
 
 | |
2FJW
 
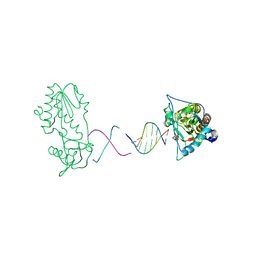 | | d(CTTGAATGCATTCAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*GP*AP*AP*TP*G)-3', 5'-D(P*CP*AP*TP*TP*CP*AP*AP*G)-3', Reverse transcriptase | | Authors: | Goodwin, K.D, Georgiadis, M.M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A High-Throughput, High-Resolution Strategy for the Study of Site-Selective DNA Binding Agents: Analysis of a "Highly Twisted" Benzimidazole-Diamidine.
J.Am.Chem.Soc., 128, 2006
|
|
5NUS
 
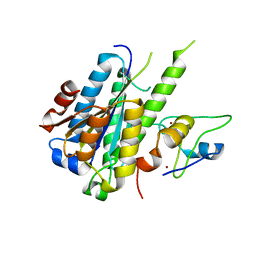 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | Descriptor: | ZINC ION, p34, p44 | | Authors: | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | Deposit date: | 2017-05-02 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
