8CPD
 
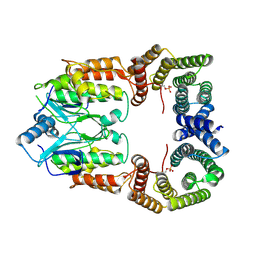 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | Descriptor: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Dedden, D, Ulrich, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8BZO
 
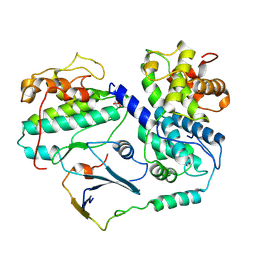 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8BYL
 
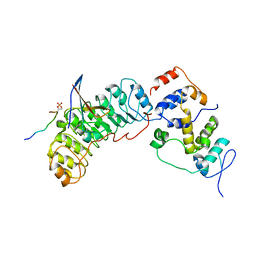 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
1AFC
 
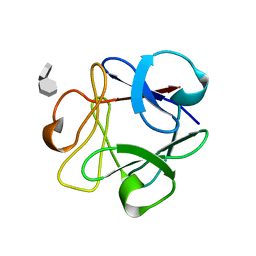 | | STRUCTURAL STUDIES OF THE BINDING OF THE ANTI-ULCER DRUG SUCROSE OCTASULFATE TO ACIDIC FIBROBLAST GROWTH FACTOR | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Hsu, B.T, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the binding of the anti-ulcer drug sucrose octasulfate to acidic fibroblast growth factor.
Structure, 1, 1993
|
|
7NIZ
 
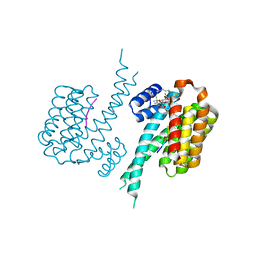 | | Human 14-3-3 sigma in complex with human Estrogen Receptor alpha peptide and ligands Fusicoccin-A and WR-1065 | | Descriptor: | 14-3-3 protein sigma, Estrogen receptor, FUSICOCCIN, ... | | Authors: | Roversi, P, Falcicchio, M, Doveston, R. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cooperative stabilisation of 14-3-3 sigma protein-protein interactions via covalent protein modification.
Chem Sci, 12, 2021
|
|
1AJY
 
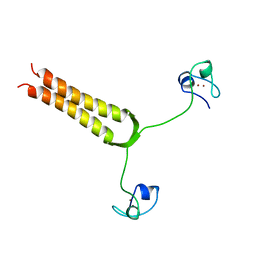 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
8CDN
 
 | | Crystal structure of human Brachyury in complex with a single T box binding element DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*TP*CP*AP*CP*AP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*TP*GP*TP*GP*AP*GP*CP*CP*T)-3'), T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, von Delft, F, Gileadi, O, Bountra, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of human Brachyury in complex with a single T box binding element DNA
To Be Published
|
|
7NND
 
 | |
7NMX
 
 | |
7NPG
 
 | |
7NNE
 
 | |
7NMW
 
 | |
7NMA
 
 | |
7NPB
 
 | |
7NP2
 
 | |
7NN2
 
 | |
1AD6
 
 | | DOMAIN A OF HUMAN RETINOBLASTOMA TUMOR SUPPRESSOR | | Descriptor: | RETINOBLASTOMA TUMOR SUPPRESSOR | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 1997-02-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box.
Nat.Struct.Biol., 4, 1997
|
|
7N16
 
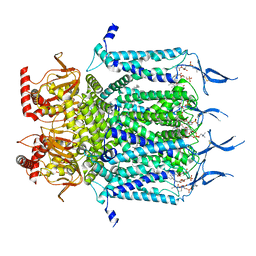 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N17
 
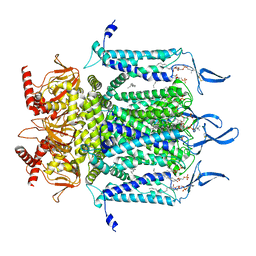 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N15
 
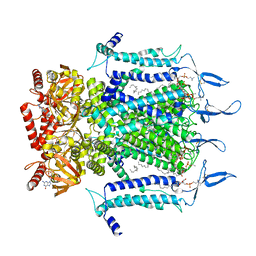 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
8CO6
 
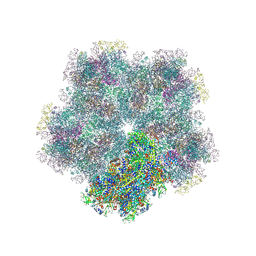 | | Subtomogram average of Immature Rotavirus TLP penton | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8C3C
 
 | |
8C2G
 
 | |
2JUL
 
 | | NMR Structure of DREAM | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Ames, J. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization.
Biochemistry, 47, 2008
|
|
