6ALF
 
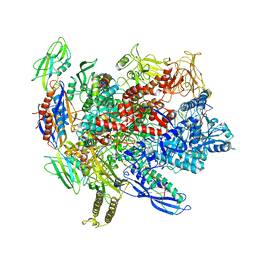 | | CryoEM structure of crosslinked E.coli RNA polymerase elongation complex | | Descriptor: | DNA (29-MER), DNA (5'-D(*GP*GP*GP*CP*TP*AP*AP*TP*GP*AP*CP*GP*GP*CP*GP*AP*AP*TP*AP*CP*CP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Darst, S.A. | | Deposit date: | 2017-08-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription arrest by coliphage HK022 Nun in anEscherichia coliRNA polymerase elongation complex.
Elife, 6, 2017
|
|
5ZVJ
 
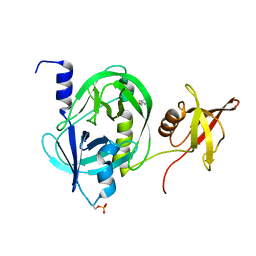 | | Crystal structure of HtrA1 from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine protease | | Authors: | Khundrakpam, H.S, Yadav, S, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-10 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an essential high-temperature requirement protein HtrA1 (Rv1223) from Mycobacterium tuberculosis reveals its unique features.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6AHV
 
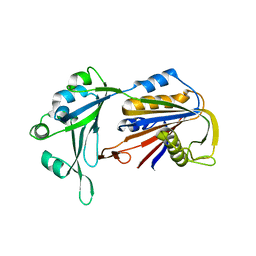 | | Crystal structure of human RPP40 | | Descriptor: | Ribonuclease P protein subunit p40 | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6ALH
 
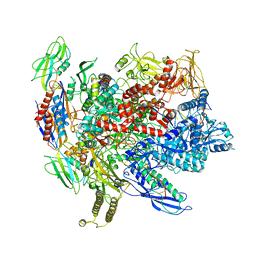 | | CryoEM structure of E.coli RNA polymerase elongation complex | | Descriptor: | DNA (29-MER), DNA (5'-D(*GP*GP*GP*CP*TP*AP*AP*TP*GP*AP*CP*GP*GP*CP*GP*AP*AP*TP*AP*CP*CP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Darst, S.A. | | Deposit date: | 2017-08-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of transcription arrest by coliphage HK022 Nun in anEscherichia coliRNA polymerase elongation complex.
Elife, 6, 2017
|
|
6AHR
 
 | | Cryo-EM structure of human Ribonuclease P | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
3CW4
 
 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
3C3B
 
 | | Crystal Structure of human phosphoglycerate kinase bound to D-CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, Phosphoglycerate kinase 1 | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
3C3A
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
2PRG
 
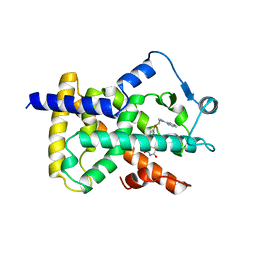 | | LIGAND-BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), NUCLEAR RECEPTOR COACTIVATOR SRC-1, ... | | Authors: | Nolte, R.T, Wisely, G.B, Milburn, M.V. | | Deposit date: | 1998-08-14 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma.
Nature, 395, 1998
|
|
2M4Z
 
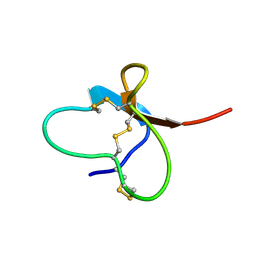 | |
2PWD
 
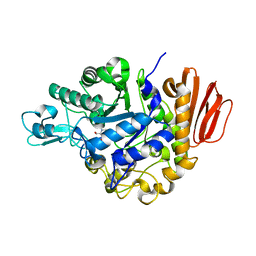 | | Crystal Structure of the Trehalulose Synthase MUTB from Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Deoxynojirmycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWF
 
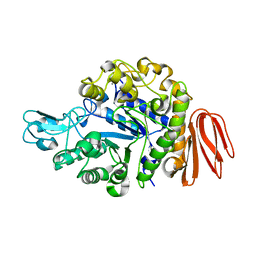 | | Crystal structure of the MutB D200A mutant in complex with glucose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
3BEH
 
 | |
3BDL
 
 | | Crystal structure of a truncated human Tudor-SN | | Descriptor: | CITRIC ACID, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Li, C.L. | | Deposit date: | 2007-11-15 | | Release date: | 2008-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing.
Nucleic Acids Res., 36, 2008
|
|
2MRE
 
 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2MVZ
 
 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
2PWH
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2RK2
 
 | | DHFR R-67 complexed with NADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
2LER
 
 | | Conotoxin pc16a | | Descriptor: | Conotoxin pc16a | | Authors: | Dyubankova, N, Lescrinier, E, Van Der Haegen, A, Peigneur, S, Tytgat, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-04-11 | | Last modified: | 2012-04-18 | | Method: | SOLUTION NMR | | Cite: | Pc16a, the first characterized peptide from Conus pictus venom, shows a novel disulfide connectivity.
Peptides, 34, 2012
|
|
2LSU
 
 | | The NMR high resolution structure of yeast Tah1 in a free form | | Descriptor: | TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
2N1G
 
 | |
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2MRF
 
 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2PWE
 
 | | Crystal structure of the MutB E254Q mutant in complex with the substrate sucrose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
6BM8
 
 | | Crystal structure of glycoprotein B from Herpes Simplex Virus type I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, ... | | Authors: | Cooper, R.S, Heldwein, E.E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB.
Nat. Struct. Mol. Biol., 25, 2018
|
|
