1K6H
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1V4Q
 
 | |
1LC6
 
 | | Solution Structure of the U6 Intramolecular Stem-loop RNA | | Descriptor: | U6 Intramolecular Stem-loop RNA | | Authors: | Huppler, A, Nikstad, L.J, Allmann, A.M, Brow, D.A, Butcher, S.E. | | Deposit date: | 2002-04-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure.
Nat.Struct.Biol., 9, 2002
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1KV4
 
 | | Solution structure of antibacterial peptide (Moricin) | | Descriptor: | moricin | | Authors: | Hemmi, H, Ishibashi, J, Hara, S, Yamakawa, M. | | Deposit date: | 2002-01-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
FEBS Lett., 518, 2002
|
|
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
1QP6
 
 | | SOLUTION STRUCTURE OF ALPHA2D | | Descriptor: | PROTEIN (ALPHA2D) | | Authors: | Hill, R.B, DeGrado, W.F. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha2D, A Nativelike De Novo Designed Protein
J.Am.Chem.Soc., 120, 1998
|
|
1Q8K
 
 | |
1VAZ
 
 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1OW8
 
 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1L1P
 
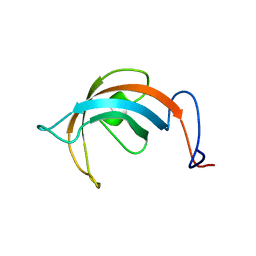 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1OWL
 
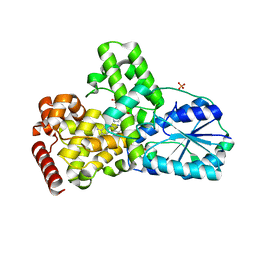 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
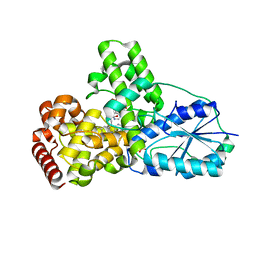 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1L6H
 
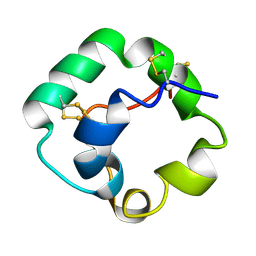 | |
1V89
 
 | | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein | | Descriptor: | Hypothetical protein KIAA0053 | | Authors: | Li, H, Tochio, N, Koshiba, S, Inoue, M, Hirota, H, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein
To be Published
|
|
1OPZ
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonneli, L, Su, X.C. | | Deposit date: | 2003-03-06 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis.
J.Mol.Biol., 331, 2003
|
|
1QA9
 
 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | Descriptor: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | Authors: | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | Deposit date: | 1999-04-13 | | Release date: | 1999-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
1QPG
 
 | | 3-PHOSPHOGLYCERATE KINASE, MUTATION R65Q | | Descriptor: | 3-PHOSPHOGLYCERATE KINASE, 3-PHOSPHOGLYCERIC ACID, MAGNESIUM-5'-ADENYLY-IMIDO-TRIPHOSPHATE | | Authors: | Mcphillips, T.M, Hsu, B.T, Sherman, M.A, Mas, M.T, Rees, D.C. | | Deposit date: | 1996-01-04 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the R65Q mutant of yeast 3-phosphoglycerate kinase complexed with Mg-AMP-PNP and 3-phospho-D-glycerate.
Biochemistry, 35, 1996
|
|
1QV6
 
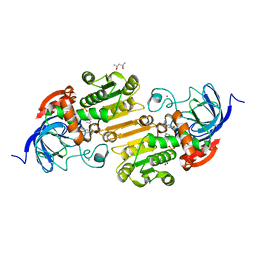 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,4-DIFLUOROBENZYL ALCOHOL | | Descriptor: | (2,4-DIFLUOROPHENYL)METHANOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alcohol dehydrogenase E chain, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-26 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
1KMG
 
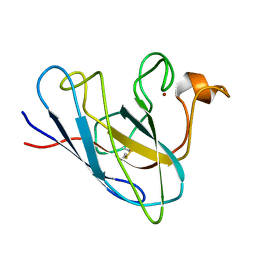 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
1TIJ
 
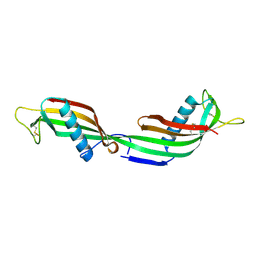 | | 3D Domain-swapped human cystatin C with amyloid-like intermolecular beta-sheets | | Descriptor: | Cystatin C | | Authors: | Janowski, R, Kozak, M, Abrahamson, M, Grubb, A, Jaskolski, M. | | Deposit date: | 2004-06-02 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | 3D domain-swapped human cystatin C with amyloidlike intermolecular beta-sheets.
Proteins, 61, 2005
|
|
1Q68
 
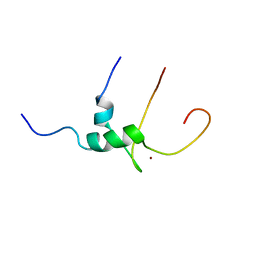 | | Solution structure of T-cell surface glycoprotein CD4 and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD4, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
