1EXD
 
 | | CRYSTAL STRUCTURE OF A TIGHT-BINDING GLUTAMINE TRNA BOUND TO GLUTAMINE AMINOACYL TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMINE TRNA APTAMER, GLUTAMINYL-TRNA SYNTHETASE, ... | | Authors: | Bullock, T.L, Sherlin, L.D, Perona, J.J. | | Deposit date: | 2000-05-02 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary core rearrangements in a tight binding transfer RNA aptamer.
Nat.Struct.Biol., 7, 2000
|
|
1YDZ
 
 | | T-To-T(High) quaternary transitions in human hemoglobin: alphaY140F oxy (2MM IHP, 20% PEG) (1 test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, OXYGEN MOLECULE, ... | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-12-27 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
4IFD
 
 | |
7Y5G
 
 | | Cryo-EM structure of a eukaryotic ZnT8 in the presence of zinc | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
1HSS
 
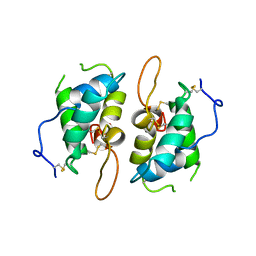 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
1K6O
 
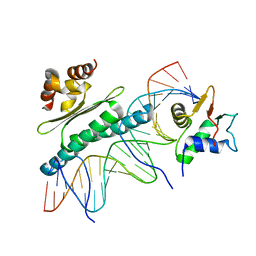 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
4FW8
 
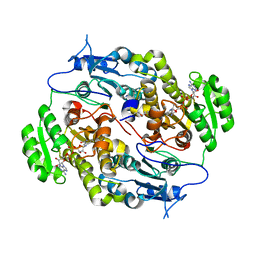 | | Crystal structure of FABG4 complexed with Coenzyme NADH | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2012-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
3GQC
 
 | | Structure of human Rev1-DNA-dNTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Swan, M.K, Aggarwal, A.K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the human Rev1-DNA-dNTP ternary complex.
J.Mol.Biol., 390, 2009
|
|
2X66
 
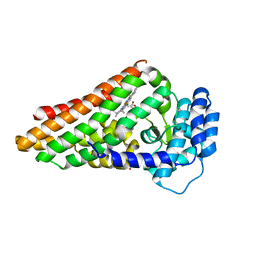 | | The binary complex of PrnB (the second enzyme in pyrrolnitrin biosynthesis pathway) and cyanide | | Descriptor: | CYANIDE ION, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhu, X, van pee, K.-H, Naismith, J.H. | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Ternary Complex of Prnb (the Second Enzyme in Pyrrolnitrin Biosynthesis Pathway), Tryptophan and Cyanide Yields New Mechanistic Insights Into the Indolamine Dioxygenase Superfamily.
J.Biol.Chem., 285, 2010
|
|
4I40
 
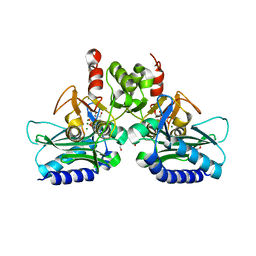 | | crystal structure of Staphylococcal inositol monophosphatase-1: 50mM LiCl inhibited complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
4I3Y
 
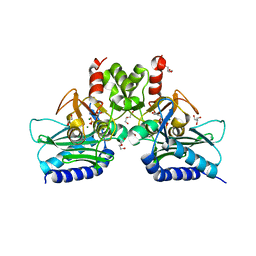 | | Crystal structure of Staphylococcal inositol monophosphatase-1: 100 mM LiCl soaked inhibitory complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
5X8X
 
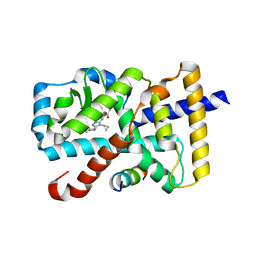 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Compound A. | | Descriptor: | (3R,4R)-4-[4-cyclopropyl-5-[3-(2-methylpropyl)cyclobutyl]-1,2,4-triazol-3-yl]-N-(2,4-dimethylphenyl)-1-ethanoyl-pyrrolidine-3-carboxamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
6DFR
 
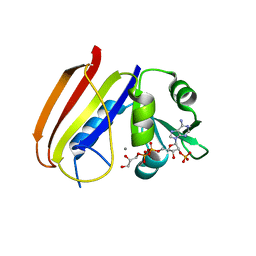 | |
8QLP
 
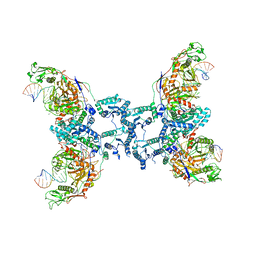 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
8QLO
 
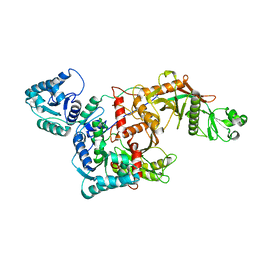 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
7TE0
 
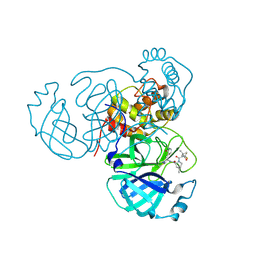 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary and Structural Insights about Potential SARS-CoV-2 Evasion of Nirmatrelvir.
J.Med.Chem., 65, 2022
|
|
7TLX
 
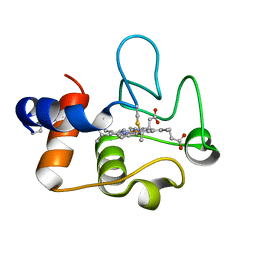 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
2CSC
 
 | |
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
2X67
 
 | | The ternary complex of PrnB (the second enzyme in pyrrolnitrin biosynthesis pathway), tryptophan and cyanide | | Descriptor: | CYANIDE ION, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhu, X, van pee, K.-H, Naismith, J.H. | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Ternary Complex of Prnb (the Second Enzyme in Pyrrolnitrin Biosynthesis Pathway), Tryptophan and Cyanide Yields New Mechanistic Insights Into the Indolamine Dioxygenase Superfamily.
J.Biol.Chem., 285, 2010
|
|
2X68
 
 | | The ternary complex of PrnB (the second enzyme in pyrrolnitrin biosynthesis pathway), 7-Cl-L-tryptophan and cyanide | | Descriptor: | 7-CHLOROTRYPTOPHAN, CYANIDE ION, PRNB, ... | | Authors: | Zhu, X, van pee, K.-H, Naismith, J.H. | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Ternary Complex of Prnb (the Second Enzyme in Pyrrolnitrin Biosynthesis Pathway), Tryptophan and Cyanide Yields New Mechanistic Insights Into the Indolamine Dioxygenase Superfamily.
J.Biol.Chem., 285, 2010
|
|
4KI5
 
 | | Cystal structure of human factor VIII C2 domain in a ternary complex with murine inhbitory antibodies 3E6 and G99 | | Descriptor: | Coagulation factor VIII, MURINE MONOCLONAL 3E6 FAB HEAVY CHAIN, MURINE MONOCLONAL 3E6 FAB LIGHT CHAIN, ... | | Authors: | Walter, J.D, Meeks, S.L, Healey, J.F, Lollar, P, Spiegel, P.C. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of the factor VIII C2 domain in a ternary complex with 2 inhibitor antibodies reveals classical and nonclassical epitopes.
Blood, 122, 2013
|
|
4DRW
 
 | | Crystal Structure of the Ternary Complex between S100A10, an Annexin A2 N-terminal Peptide and an AHNAK Peptide | | Descriptor: | Neuroblast differentiation-associated protein AHNAK, Protein S100-A10/Annexin A2 chimeric protein | | Authors: | Rezvanpour, A, Lee, T.-W, Junop, M.S, Shaw, G.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an asymmetric ternary protein complex provides insight for membrane interaction.
Structure, 20, 2012
|
|
8K88
 
 | | Structure of procaryotic Ago | | Descriptor: | DNA (41-mer), DNA/RNA (21-mer), MAGNESIUM ION, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleic acid-induced NADase activation of a short Sir2-associated prokaryotic Argonaute system.
Cell Rep, 43, 2024
|
|
8K87
 
 | | Dimer structure of procaryotic Ago | | Descriptor: | DNA (41-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nucleic acid-induced NADase activation of a short Sir2-associated prokaryotic Argonaute system.
Cell Rep, 43, 2024
|
|
