5AJN
 
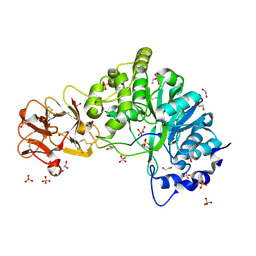 | | Crystal structure of the inactive form of GalNAc-T2 in complex with the glycopeptide MUC5AC-Cys13 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUCIN, ... | | Authors: | Lira-Navarrete, E, delasRivas, M, Companon, I, Pallares, M.C, Kong, Y, Iglesias-Fernandez, J, Bernardes, G.J.L, Peregrina, J.M, Rovira, C, Bernado, P, Bruscolini, P, Clausen, H, Lostao, A, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Nat.Commun., 6, 2015
|
|
5ADC
 
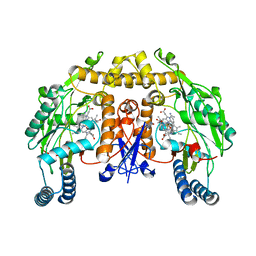 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(((5-((Methylamino)methyl)pyridin-3-yl)oxy)methyl) quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[5-[(methylideneamino)methyl]pyridin-3-yl]oxymethyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4ZS7
 
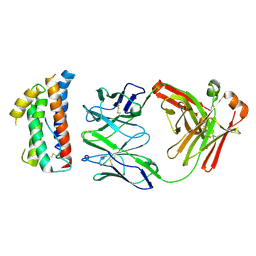 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
7N96
 
 | |
7N8D
 
 | |
7N57
 
 | |
7N4O
 
 | |
7N8G
 
 | | Crystal structure of R20A-R22A human Galectin-7 mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2021-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of R20A human Galectin-7 mutant
To Be Published
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
7NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, ADMA COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
6GXD
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD752MS after reaction initiation | | Descriptor: | CHLORIDE ION, fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6GH7
 
 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(3~{R})-pyrrolidin-3-yl]pentanamide, Streptavidin | | Authors: | Nodling, A.R, Tsai, Y.H, Luk, L.Y.P, Rizkallah, P, Jin, Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Reactivity and Selectivity of Iminium Organocatalysis Improved by a Protein Host.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
6GXO
 
 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and P/E-tRNA (State IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GJ0
 
 | | Human IMPase with Mn | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | Authors: | Kraft, L.V, Roe, S.M. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Co-crystallization of human inositol monophosphatase with the lithium mimetic L-690,330.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7NXC
 
 | |
5DCG
 
 | |
6H0F
 
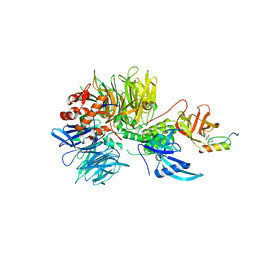 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
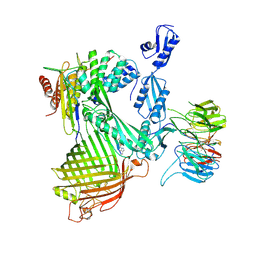 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
5DFJ
 
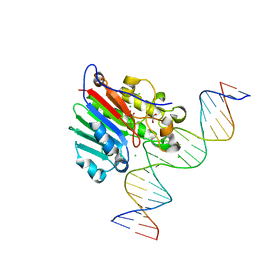 | | Human APE1 E96Q/D210N mismatch substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7NVT
 
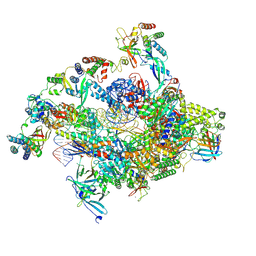 | | RNA polymerase II core pre-initiation complex with closed promoter DNA in distal position | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVU
 
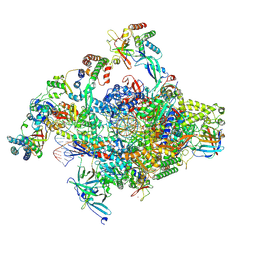 | | RNA polymerase II core pre-initiation complex with open promoter DNA | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
6EQD
 
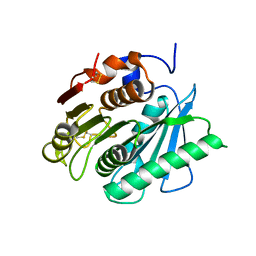 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ENJ
 
 | |
