2A63
 
 | |
8QTQ
 
 | | Thermostable WW domain | | Descriptor: | WW domain | | Authors: | Kovermann, M, Thomas, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Thermostable WW-Domain Scaffold to Design Functional beta-Sheet Miniproteins.
J.Am.Chem.Soc., 2024
|
|
1E0G
 
 | | LYSM Domain from E.coli MLTD | | Descriptor: | Membrane-bound lytic murein transglycosylase D | | Authors: | Bateman, A, Bycroft, M. | | Deposit date: | 2000-03-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of a Lysm Domain from E.Coli Membrane Bound Lytic Murein Transglycosylase D (Mltd)
J.Mol.Biol., 299, 2000
|
|
7OQT
 
 | |
7OSR
 
 | |
7OSW
 
 | |
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1DVV
 
 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
1DXN
 
 | |
8R1X
 
 | |
1Z2D
 
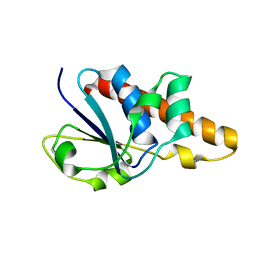 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
8OH7
 
 | |
1X6B
 
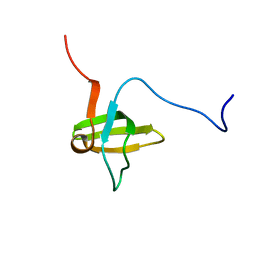 | | Solution structures of the SH3 domain of human rho guanine exchange factor (GEF) 16 | | Descriptor: | Rho guanine exchange factor (GEF) 16 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domain of human rho guanine exchange factor (GEF) 16
To be Published
|
|
1X6H
 
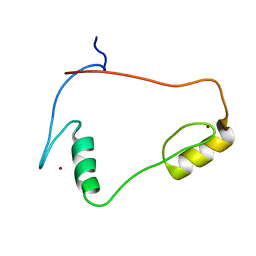 | | Solution structures of the C2H2 type zinc finger domain of human Transcriptional repressor CTCF | | Descriptor: | Transcriptional repressor CTCF, ZINC ION | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C2H2 type zinc finger domain of human Transcriptional repressor CTCF
To be Published
|
|
1X63
 
 | |
1X6G
 
 | | Solution structures of the SH3 domain of human megakaryocyte-associated tyrosine-protein kinase. | | Descriptor: | Megakaryocyte-associated tyrosine-protein kinase | | Authors: | Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domain of human megakaryocyte-associated tyrosine-protein kinase.
To be Published
|
|
1X68
 
 | | Solution structures of the C-terminal LIM domain of human FHL5 protein | | Descriptor: | FHL5 protein, ZINC ION | | Authors: | Nameki, N, Sasagawa, A, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C-terminal LIM domain of human FHL5 protein
To be Published
|
|
8SEM
 
 | |
1X6D
 
 | | Solution structures of the PDZ domain of human Interleukin-16 | | Descriptor: | Interleukin-16 | | Authors: | Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PDZ domain of human Interleukin-16
To be Published
|
|
5H3N
 
 | |
8OFC
 
 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | Descriptor: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | Authors: | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 52, 2024
|
|
8R8P
 
 | |
2AYM
 
 | |
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
1Y49
 
 | |
