7LRW
 
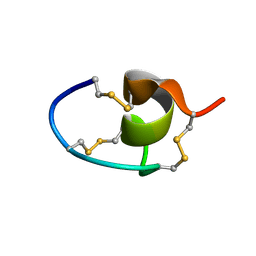 | |
7LT7
 
 | | Structure of Hact-3 | | Descriptor: | Hact-3 | | Authors: | Schmidt, C.A, Daly, N.L. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity of coral derived peptides
To Be Published
|
|
7LX4
 
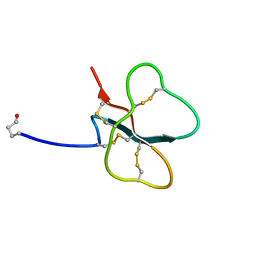 | |
6KJO
 
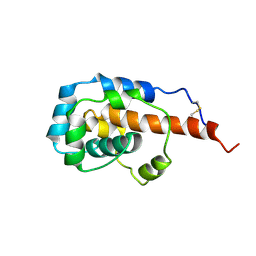 | | The microtubule-binding domains of yeast cytoplasmic dynein in the low affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6QVW
 
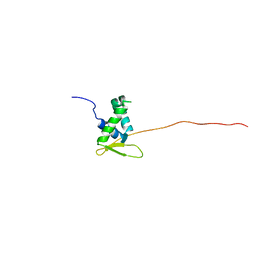 | | Solution structure of the free FOXO1 DNA binding domain | | Descriptor: | Forkhead box protein O1 | | Authors: | Psenakova, K, Obsil, T, Veverka, V, Obsilova, V, Kohoutova, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics.
Cells, 8, 2019
|
|
6K3X
 
 | |
6R14
 
 | | Structure of kiteplatinated dsDNA | | Descriptor: | Kiteplatin, Kiteplatinated DNA oligomer, chain A, ... | | Authors: | Margiotta, N, Papadia, P, Kubicek, K, Krejcikova, M, Gkionis, K, Sponer, J. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of kiteplatinated DNA
To Be Published
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1K29
 
 | | Solution Structure of a DNA Duplex Containing M1G Opposite a 2 Base Pair Deletion | | Descriptor: | 5'-D(*AP*TP*CP*GP*CP*(M1G)P*CP*GP*GP*CP*AP*TP*G)-3', 5'-D(*CP*AP*TP*GP*CP*CP*GP*CP*GP*AP*T)-3' | | Authors: | Schnetz-Boutaud, N.C, Saleh, S, Marnett, L.J, Stone, M.P. | | Deposit date: | 2001-09-26 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The exocyclic 1,N2-deoxyguanosine pyrimidopurinone M1G is a chemically stable DNA adduct when placed opposite a two-base deletion in the (CpG)3 frameshift hotspot of the Salmonella typhimurium hisD3052 gene.
Biochemistry, 40, 2001
|
|
6K84
 
 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
6R9K
 
 | |
6R9L
 
 | |
6QTC
 
 | | tSH2 domain of transcription elongation factor Spt6 complexed with tyrosine phosphorylated CTD | | Descriptor: | Tyrosine phosphorylated CTD, tSH2 domain of transcription elongation factor Spt6 | | Authors: | Brazda, P, Krejcikova, M, Smirakova, E, Kubicek, K, Stefl, R. | | Deposit date: | 2019-02-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | tSH2 domain of transcription elongation factor Spt6 complexed with tyrosine phosphorylated CTD
To Be Published
|
|
1IB9
 
 | |
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
7MJ3
 
 | | Structure of Hact-4 | | Descriptor: | Hact-4 | | Authors: | Schmidt, C.A, Daly, N.L. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diversity of coral derived peptides
To Be Published
|
|
7M1D
 
 | |
7NIP
 
 | |
6KXZ
 
 | |
1HWQ
 
 | |
1HY8
 
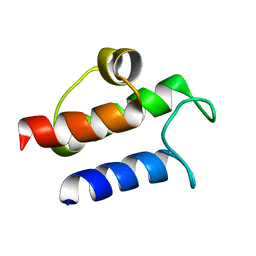 | | SOLUTION STRUCTURE OF B. SUBTILIS ACYL CARRIER PROTEIN | | Descriptor: | ACYL CARRIER PROTEIN | | Authors: | Xu, G.-Y, Tam, A, Lin, L, Hixon, J, Fritz, C.C, Power, R. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of B. subtilis acyl carrier protein.
Structure, 9, 2001
|
|
1HWV
 
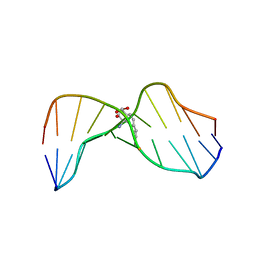 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1CXW
 
 | | THE SECOND TYPE II MODULE FROM HUMAN MATRIX METALLOPROTEINASE 2 | | Descriptor: | HUMAN MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Grishaev, A, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-12 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | The second type II module from human matrix metalloproteinase 2: structure, function and dynamics.
Structure Fold.Des., 7, 1999
|
|
6Y4O
 
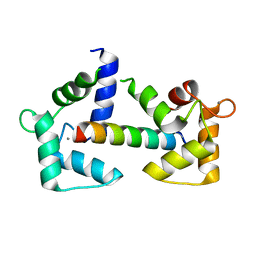 | | Calmodulin bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-2, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83549082 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
