9C8T
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with AMPPNP and compound 2 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, GLYCEROL, ... | | Authors: | Wang, L, Sietsema, D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural insight into the cGAS active site explains differences between therapeutically relevant species.
Commun Chem, 8, 2025
|
|
9JEU
 
 | | Crystal structure of a cupin protein (tm1459) in iron (Fe) substituted form | | Descriptor: | Cupin type-2 domain-containing protein, FE (III) ION | | Authors: | Fujieda, N, Ichihashi, H, Kurisu, G, Itoh, S. | | Deposit date: | 2024-09-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Unusual Self-Hydroxylation in 4-Histidine Tetrad-Supporting Nonheme Iron Center.
Chem Asian J, 20, 2025
|
|
9C8N
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with AMPPNP and compound 1 | | Descriptor: | 1-[(1S)-6,7-dichloro-1-methyl-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-methoxyethan-1-one, Cyclic GMP-AMP synthase, GLYCEROL, ... | | Authors: | Wang, L, Sietsema, D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the cGAS active site explains differences between therapeutically relevant species.
Commun Chem, 8, 2025
|
|
8ASE
 
 | | Crystal structure of Thrombin in complex with macrocycle T3 | | Descriptor: | (8~{S},14~{S},18~{E})-8-[(4-chlorophenyl)methyl]-3,21-dithia-7,10,16-triazatricyclo[21.2.2.1^{10,14}]octacosa-1(26),18,23(27),24-tetraene-6,9,15-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chinellato, M, Angelini, A, Nielsen, A, Heinis, C, Cendron, L. | | Deposit date: | 2022-08-19 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thrombin in complex with optimized macrocycles T1 and T3
To Be Published
|
|
4ZA6
 
 | |
9CS1
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP2 | | Descriptor: | Cyclic peptide CP2, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
4OGQ
 
 | | Internal Lipid Architecture of the Hetero-Oligomeric Cytochrome b6f Complex | | Descriptor: | (1R)-1-{[(2S)-3-hydroxy-2-{[(1R)-1-hydroxypentyl]oxy}propyl]oxy}hexan-1-ol, (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1S,8E)-1-{[(2S)-1-hydroxy-3-{[(1S)-1-hydroxypentadecyl]oxy}propan-2-yl]oxy}heptadec-8-en-1-ol, ... | | Authors: | Hasan, S.S, Cramer, W.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Internal Lipid Architecture of the Hetero-Oligomeric Cytochrome b6f Complex.
Structure, 22, 2014
|
|
5TA8
 
 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6S26
 
 | | Crystal structure of human wild type STING in complex with 2'-3'-cyclic-GMP-7-deaza-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M, Brynda, J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic Preparation of 2'-5',3'-5'-Cyclic Dinucleotides, Their Binding Properties to Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations.
J.Med.Chem., 62, 2019
|
|
5S20
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
9JET
 
 | | Crystal structure of a cupin protein (tm1459) in manganese (Mn) substituted form | | Descriptor: | Cupin type-2 domain-containing protein, MANGANESE (II) ION | | Authors: | Fujieda, N, Ichihashi, H, Kurisu, G, Itoh, S. | | Deposit date: | 2024-09-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Unusual Self-Hydroxylation in 4-Histidine Tetrad-Supporting Nonheme Iron Center.
Chem Asian J, 20, 2025
|
|
6S38
 
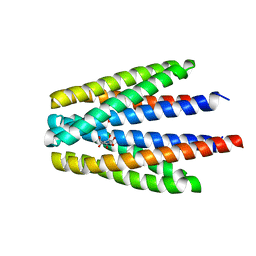 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
5Z85
 
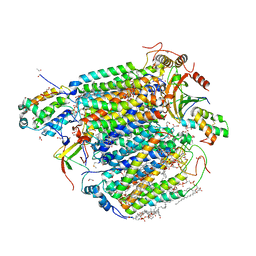 | | The structure of azide-bound cytochrome c oxidase determined using the another batch crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5S2R
 
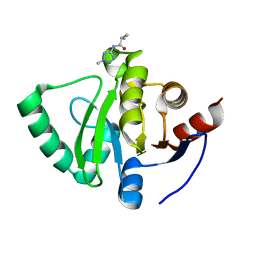 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57292369 | | Descriptor: | 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.132 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4C
 
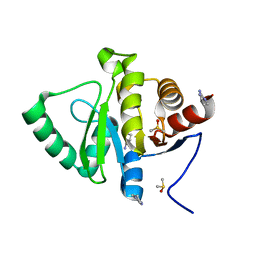 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1954800348 | | Descriptor: | 1,4,5,6-tetrahydropyrimidin-2-amine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3MYK
 
 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, MAGNESIUM ION, Myosin-2 heavy chain, ... | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
5S8Q
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with FMO3D000185a (space group P212121) | | Descriptor: | (3R)-3-methyl-1,2,3,4-tetrahydro-5H-1,4-benzodiazepin-5-one, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XChem group deposition
To Be Published
|
|
9J2Y
 
 | | Human cGAS catalytic domain bound with G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-08-07 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of novel cGAS inhibitors
To Be Published
|
|
5H2J
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5ZQO
 
 | | Tankyrase-2 in complex with compound 1a | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5S3S
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0103 | | Descriptor: | 1-[(5S,8R)-6,7,8,9-tetrahydro-5H-5,8-epiminocyclohepta[b]pyridin-10-yl]ethan-1-one, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5H2P
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6IBM
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfate ME776 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
5Z84
 
 | | The structure of azide-bound cytochrome c oxidase determined using the crystals exposed to 20 mM azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
6IIU
 
 | | Crystal structure of the human thromboxane A2 receptor bound to ramatroban | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[(3R)-3-[(4-fluorophenyl)sulfonylamino]-1,2,3,4-tetrahydrocarbazol-9-yl]propanoic acid, CHOLESTEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
