6QFD
 
 | | The complex structure of hsRosR-S4 (vng0258/RosR-S4) | | Descriptor: | DNA (28-MER), DNA-binding protein, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QFW
 
 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 9) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[3-(8-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8$l^{4}-diaza-9$l^{7}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)propyl]benzenesulfonamide, Carbonic anhydrase 2, IRIDIUM ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
1JEK
 
 | | Visna TM CORE STRUCTURE | | Descriptor: | ENV POLYPROTEIN | | Authors: | Malashkevich, V.N, Singh, M, Kim, P.S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The trimer-of-hairpins motif in membrane fusion: Visna virus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7TBG
 
 | | AtTPC1 D454N with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, SODIUM ION, Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-22 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QD7
 
 | | EM structure of a EBOV-GP bound to 3T0331 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV.
Nat. Med., 25, 2019
|
|
7SPC
 
 | | Models for C17 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208). | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
1JEO
 
 | | Crystal Structure of the Hypothetical Protein MJ1247 from Methanococcus jannaschii at 2.0 A Resolution Infers a Molecular Function of 3-Hexulose-6-Phosphate isomerase. | | Descriptor: | CITRIC ACID, HYPOTHETICAL PROTEIN MJ1247 | | Authors: | Martinez-Cruz, L.A, Dreyer, M.K, Boisvert, D.C, Yokota, H, Martinez-Chantar, M.L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-06-18 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase.
Structure, 10, 2002
|
|
7TD2
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
1JEV
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KWK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS TRP LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
6QDZ
 
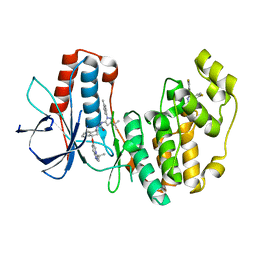 | | P38 alpha complex with AR117045 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{S})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P38 alpha complex with AR117045
To Be Published
|
|
1JF6
 
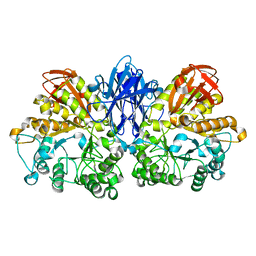 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
7STZ
 
 | |
6QG4
 
 | |
1JFS
 
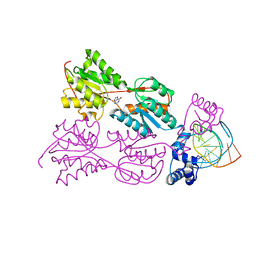 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
6QG6
 
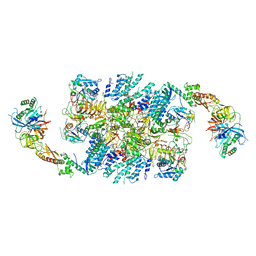 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model D) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
7TDM
 
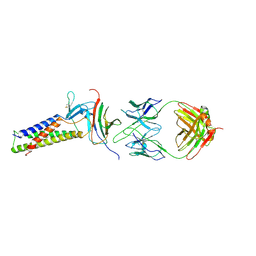 | |
7T57
 
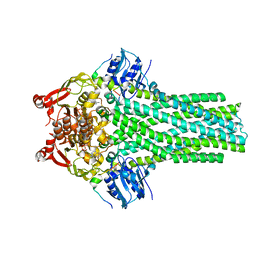 | |
1JG1
 
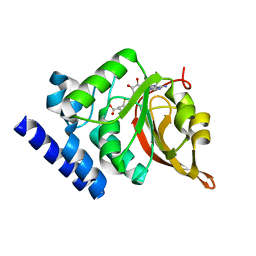 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
6QH1
 
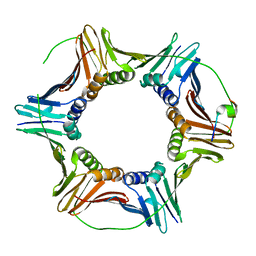 | | The structure of Schizosaccharomyces pombe PCNA in complex with an Spd1 derived peptide | | Descriptor: | Proliferating cell nuclear antigen, S-phase delaying protein 1 | | Authors: | Kragelund, B.B, Nielsen, O, Olsen, J.G, Kassem, N, Prestel, A. | | Deposit date: | 2019-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Schizosaccharomyces pombe PCNA in complex with an Spd1 derived peptide
To Be Published
|
|
7TD3
 
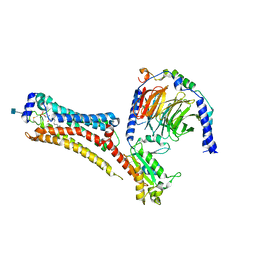 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
6QE9
 
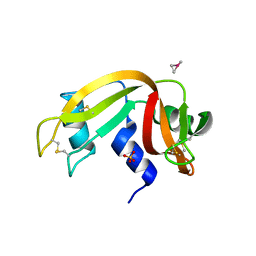 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, pentacoordinate Pt(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
7TD1
 
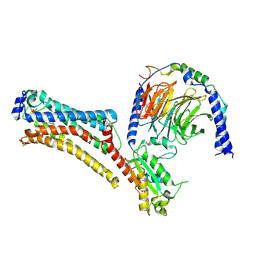 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
6QEJ
 
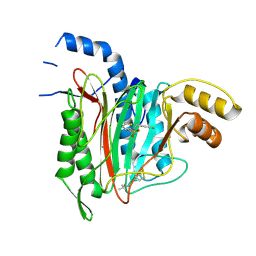 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR Thiophene-2-sulfonic acid (4-fluoro-benzyl)-(4H-[1,2,4]triazol-3-ylmethyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QHT
 
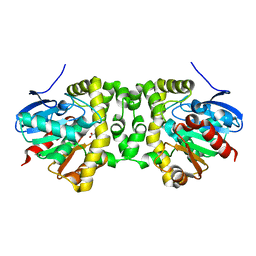 | | Time resolved structural analysis of the full turnover of an enzyme - 376 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
7TBZ
 
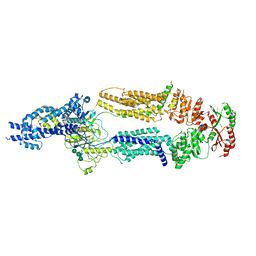 | | The structure of ABCA1 Y482C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
