5XOE
 
 | |
2DXD
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Nucleoside diphosphate kinase | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog
To be Published
|
|
5ZBJ
 
 | |
4EFC
 
 | | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Wernimont, A.K, Loppnau, P, Osman, K.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Robinson, D.A, Wyatt, P.G, Gilbert, I.H, Fairlamb, A.H, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560
To be Published
|
|
2GPB
 
 | |
2ZR2
 
 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
4H36
 
 | | Crystal Structure of JNK3 in Complex with ATF2 Peptide | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 10 | | Authors: | Nwachukwu, J.C, Laughlin, J.D, Figuera-Losada, M, Cherry, L, Nettles, K.W, LoGrasso, P.V. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanisms of Allostery and Autoinhibition in JNK Family Kinases.
Structure, 20, 2012
|
|
1CNQ
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-6-PHOSPHATE AND ZINC IONS | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, PHOSPHATE ION, ... | | Authors: | Choe, J, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-05-21 | | Release date: | 1999-05-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 37, 1998
|
|
4V1U
 
 | |
4ZKL
 
 | | Crystal structure of human histidine triad nucleotide-binding protein 1 (hHINT1) complexed with JB419 (AP4A analog) | | Descriptor: | (2R,3R,4S,5R)-2-(6-aminopurin-9-yl)-5-[2-[[(2S)-3-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-sulfanyl-phosphoryl]oxyethyl]oxolane-3,4-diol, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Kaczmarek, R, Seda, A, Krakowiak, A, Baraniak, J, Nawrot, B. | | Deposit date: | 2015-04-30 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallographic studies of the complex of human HINT1 protein with a non-hydrolyzable analog of Ap4A.
Int.J.Biol.Macromol., 87, 2016
|
|
1FPB
 
 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
4W8O
 
 | | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, luciferase-like enzymeAMP-CoA-ligase | | Authors: | Santos, C.R, Prado, R.A, Viviani, V, Murakami, M.T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm
To Be Published
|
|
6EA3
 
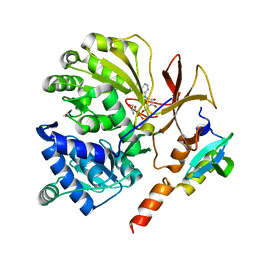 | |
2JDI
 
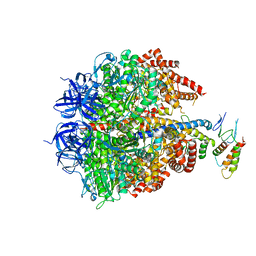 | | Ground state structure of F1-ATPase from bovine heart mitochondria (Bovine F1-ATPase crystallised in the absence of azide) | | Descriptor: | ATP SYNTHASE DELTA CHAIN, ATP SYNTHASE EPSILON CHAIN, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ground State Structure of F1-ATPase from Bovine Heart Mitochondria at 1.9 A Resolution
J.Biol.Chem., 282, 2007
|
|
4TND
 
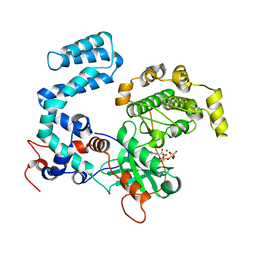 | |
5IKO
 
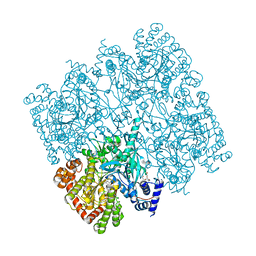 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
3NGS
 
 | | Structure of Leishmania nucleoside diphosphate kinase b with ordered nucleotide-binding loop | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Nucleoside diphosphate kinase, PHOSPHATE ION | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3NGU
 
 | | Structure of Leishmania NDKb complexed with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3NGR
 
 | | Crystal structure of Leishmania nucleoside diphosphate kinase b with unordered nucleotide-binding loop. | | Descriptor: | Nucleoside diphosphate kinase, PHOSPHATE ION | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3NDP
 
 | | Crystal structure of human AK4(L171P) | | Descriptor: | Adenylate kinase isoenzyme 4, SULFATE ION | | Authors: | Liu, R, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
6Z15
 
 | | Human wtSTING in complex with 3',3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1E24
 
 | | LYSYL-TRNA SYNTHETASE (LYSU) HEXAGONAL FORM complexed with lysine and ATP and MN2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, LYSINE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
1E1T
 
 | | LYSYL-TRNA SYNTHETASE (LYSU) HEXAGONAL FORM COMPLEXED WITH THE LYSYL_ADENYLATE INTERMEDIATE | | Descriptor: | ADENOSINE-5'-[LYSYL-PHOSPHATE], GLYCEROL, LYSYL-TRNA SYNTHETASE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-10 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
1E1O
 
 | | lysyl-tRNA Synthetase (LYSU) hexagonal form, complexed with lysine | | Descriptor: | GLYCEROL, LYSINE, LYSYL-TRNA SYNTHETASE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
4XGP
 
 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized and soaked with AMP. | | Descriptor: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, ADENINE, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2015-01-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
