5HPB
 
 | | human dihydrofolate reductase complex with NADPH and 5-methyl-6-(phenylthio-4'trifluoromethyl)thieno[2,3-d]pyrimidine-2,4-diamine | | Descriptor: | 5-methyl-6-{[4-(trifluoromethyl)phenyl]sulfanyl}thieno[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Ganjee, A. | | Deposit date: | 2016-01-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Human dihydrofolate reductase ternary complex with a series offluorine substituted 5-methyl-6-phenythio)thieno[2,3-d]pyrimidine-2,4-diamines
To Be Published
|
|
8IU1
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2R,3R,4S,5R,6R)-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-5-oxidanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-yl]oxyethanoic acid, Galectin-3, MAGNESIUM ION | | Authors: | Jinal, S, Amit, K, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
8ITZ
 
 | | Crystal structure of human Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2R,3R,4S,5R,6R)-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-5-oxidanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-yl]oxyethanoic acid, CHLORIDE ION, Galectin-3, ... | | Authors: | Jinal, S, Amit, K, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
1H88
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
3LJF
 
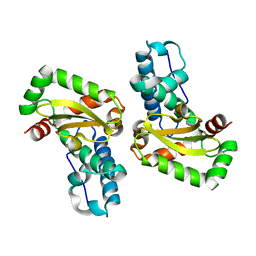 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
8ITX
 
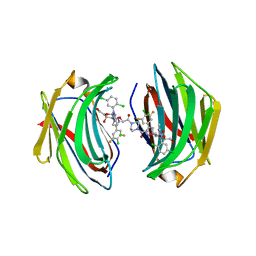 | | Crystal structure of human Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2S,3R,4S,5R,6R)-2-[2-[2,5-bis(chloranyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-5-oxidanyl-oxan-3-yl]oxyethanoic acid, CHLORIDE ION, Galectin-3, ... | | Authors: | Jinal, S, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
8J44
 
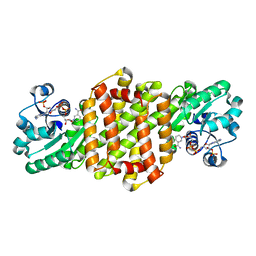 | | Reductive Aminase RA34-WT | | Descriptor: | 6-phosphogluconate dehydrogenase NAD-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
8J43
 
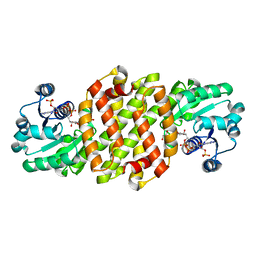 | | Reductive aminase RA29-WT | | Descriptor: | 1,2-ETHANEDIOL, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
6KZS
 
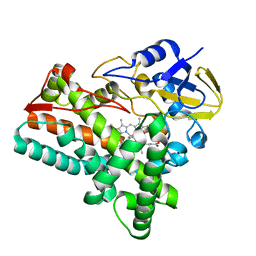 | | Crystal structure of cytochrome P450mel 107F1 in complex with heme and imidazole | | Descriptor: | Cytochrome P-450, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
8G8D
 
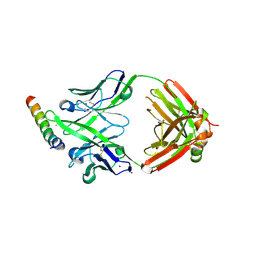 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8A
 
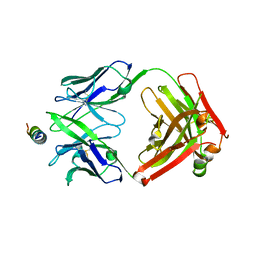 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8W0K
 
 | |
1GSH
 
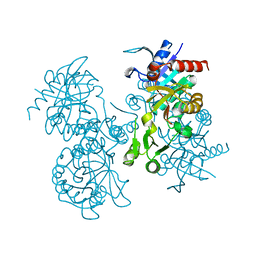 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1OYV
 
 | | Crystal structure of tomato inhibitor-II in a ternary complex with subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Subtilisin Carlsberg, Wound-induced proteinase inhibitor-II | | Authors: | Barrette-Ng, I.H, Ng, K.K, Cherney, M.M, Pearce, G, Ryan, C.A, James, M.N. | | Deposit date: | 2003-04-07 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg
J.Biol.Chem., 278, 2003
|
|
8GB0
 
 | |
1I8O
 
 | |
6ROB
 
 | | Human Carbonic Anhydrase II in complex with 4-cyanobenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-cyanobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.929 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6ZNR
 
 | | MaeB PTA domain R535A mutant | | Descriptor: | Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNU
 
 | | MaeB PTA domain E544R mutant | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNJ
 
 | | MaeB full-length enzyme apoprotein form | | Descriptor: | NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6RRG
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3,5-DIFLUOROBENZENESULFONAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.127 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6RS5
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,5,6-tetrakis(fluoranyl)-4-methyl-benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-05-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6ZN4
 
 | | MaeB malic enzyme domain apoprotein | | Descriptor: | MAGNESIUM ION, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZN7
 
 | | MaeB malic enzyme domain apoprotein | | Descriptor: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNK
 
 | | MaeB PTA domain N718D mutant | | Descriptor: | Malate dehydrogenase, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.039 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
